1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
3WE1
 
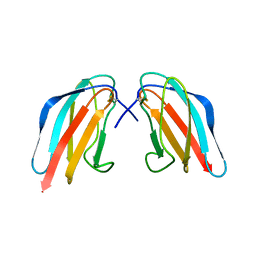 | | Crystal structure of Dengue 4 Envelope protein domain III (ED3) | | Descriptor: | Envelope protein E | | Authors: | Elahi, M, Islam, M.M, Noguchi, K, Yohda, M, Toh, H, Kuroda, Y. | | Deposit date: | 2013-06-27 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Computational prediction and experimental characterization of a "size switch type repacking" during the evolution of dengue envelope protein domain III (ED3).
Biochim.Biophys.Acta, 1844, 2014
|
|
2DRE
 
 | | Crystal structure of Water-soluble chlorophyll protein from lepidium virginicum at 2.00 angstrom resolution | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Horigome, D, Satoh, H, Itoh, N, Mitsunaga, K, Oonishi, I, Nakagawa, A, Uchida, A. | | Deposit date: | 2006-06-08 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein.
J.Biol.Chem., 282, 2007
|
|
1K1V
 
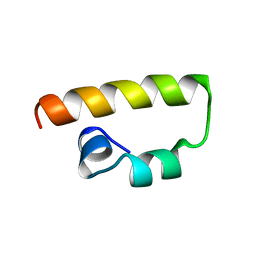 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
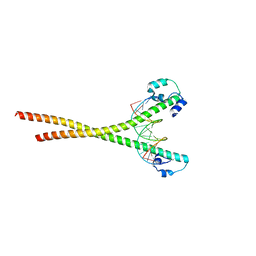 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
2LHI
 
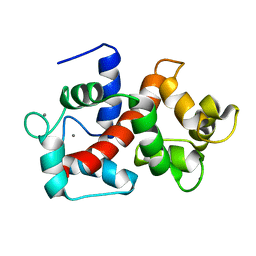 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
6GIW
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-a | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
6GIX
 
 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum (Mutation L91P) with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Palm, D.M, Agostini, A, Averesch, V, Girr, P, Werwie, M, Takahashi, S, Satoh, H, Jaenicke, E, Paulsen, H. | | Deposit date: | 2018-05-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein.
Nat Plants, 4, 2018
|
|
1WYW
 
 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
5A6W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5A6P
 
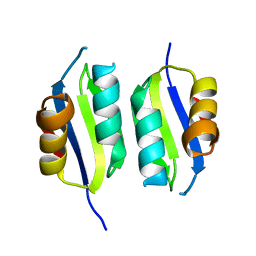 | | Heavy metal associated domain of NLR-type immune receptor Pikp1 from rice (Oryza sativa) | | Descriptor: | RESISTANCE PROTEIN PIKP-1 | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
2D07
 
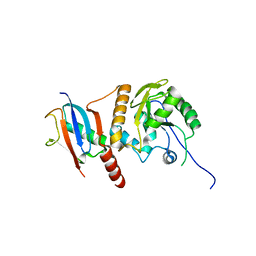 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
3ASQ
 
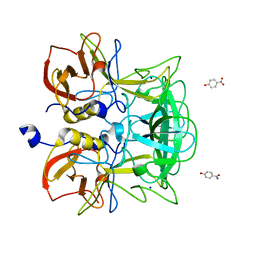 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
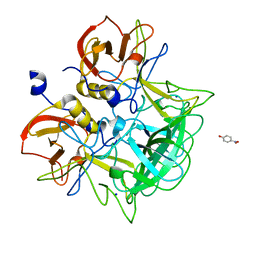 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
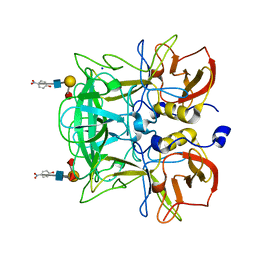 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASP
 
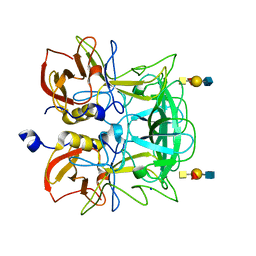 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3AST
 
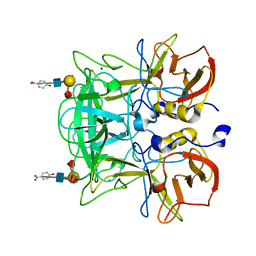 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3A5T
 
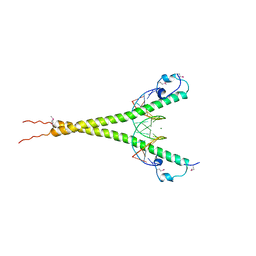 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
3A4S
 
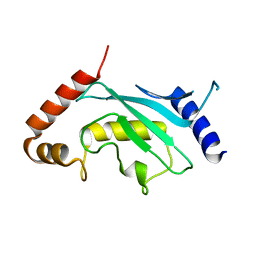 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
3A4R
 
 | | The crystal structure of SUMO-like domain 2 in Nip45 | | Descriptor: | 1,2-ETHANEDIOL, NFATC2-interacting protein, SULFATE ION | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
