5UJM
 
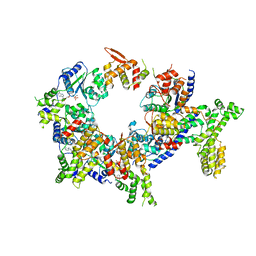 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5UJ8
 
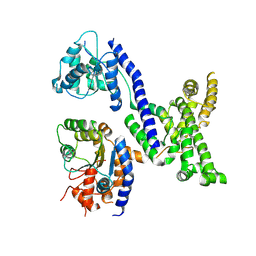 | | Human Origin Recognition Complex subunits 2 and 3 | | Descriptor: | Origin recognition complex subunit 2, Origin recognition complex subunit 3 | | Authors: | Tocilj, A, On, K.F, Elkayam, E, Joshua-Tor, L. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5UJ7
 
 | | Structure of the active form of human Origin Recognition Complex ATPase motor module, complex subunitS 1, 4, 5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, Elkayam, E, On, K.F, Joshua-Tor, L. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
1DV4
 
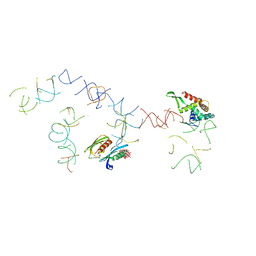 | | PARTIAL STRUCTURE OF 16S RNA OF THE SMALL RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS | | Descriptor: | 16S RIBOSOMAL RNA, OCTADECATUNGSTENYL DIPHOSPHATE, RIBOSOMAL PROTEIN S5, ... | | Authors: | Tocilj, A, Schlunzen, F, Janell, D, Gluhmann, M, Hansen, H, Harms, J, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The small ribosomal subunit from Thermus thermophilus at 4.5 A resolution: pattern fittings and the identification of a functional site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1YNI
 
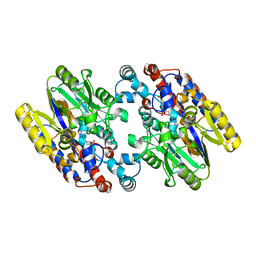 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNH
 
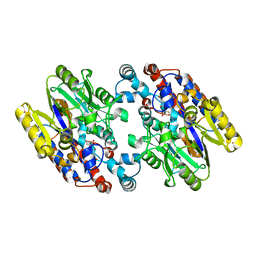 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ORNITHINE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
1YNF
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | POTASSIUM ION, Succinylarginine dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
3B8N
 
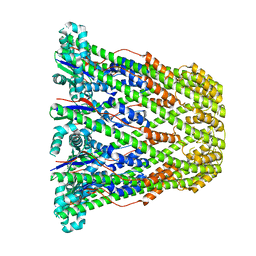 | |
3B8P
 
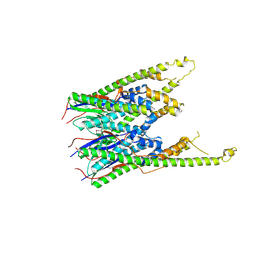 | |
3B8O
 
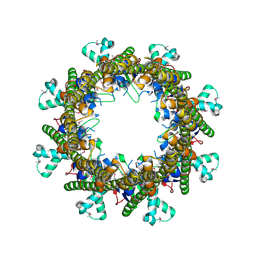 | |
3B8M
 
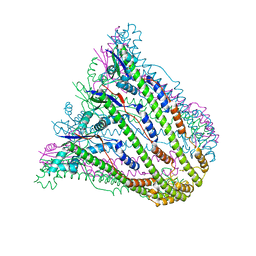 | |
4F3T
 
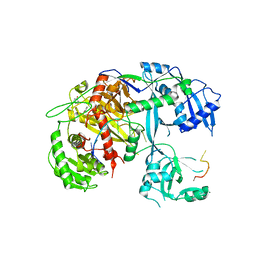 | | Human Argonaute-2 - miR-20a complex | | Descriptor: | PHENOL, Protein argonaute-2, RNA (5'-R(P*UP*AP*AP*AP*GP*UP*GP*CP*UP*UP*AP*UP*AP*GP*UP*G*CP*AP*GP*G)-3') | | Authors: | Elkayam, E, Kuhn, C.-D, Tocilj, A, Joshua-Tor, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of Human Argonaute-2 in Complex with miR-20a.
Cell(Cambridge,Mass.), 150, 2012
|
|
1VA4
 
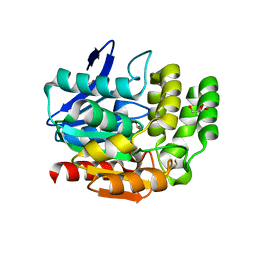 | | Pseudomonas fluorescens aryl esterase | | Descriptor: | Arylesterase, GLYCEROL | | Authors: | Cheeseman, J.D, Tocilj, A, Park, S, Schrag, J.D, Kazlauskas, R.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure of an aryl esterase from Pseudomonas fluorescens.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SBZ
 
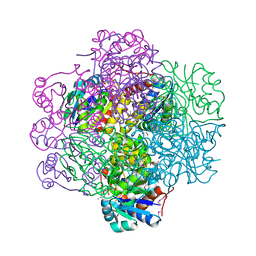 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
1P9N
 
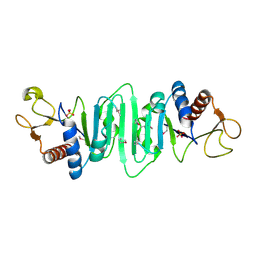 | | Crystal structure of Escherichia coli MobB. | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Rangarajan, S.E, Tocilj, A, Li, Y, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecules of Escherichia coli MobB assemble into densely packed hollow cylinders in a crystal lattice with 75% solvent content.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1EZG
 
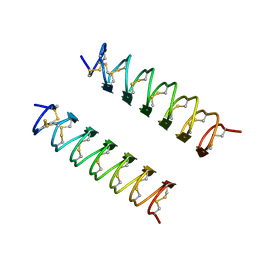 | | CRYSTAL STRUCTURE OF ANTIFREEZE PROTEIN FROM THE BEETLE, TENEBRIO MOLITOR | | Descriptor: | THERMAL HYSTERESIS PROTEIN ISOFORM YL-1 | | Authors: | Liou, Y.-C, Tocilj, A, Davies, P.L, Jia, Z. | | Deposit date: | 2000-05-10 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein.
Nature, 406, 2000
|
|
2IMT
 
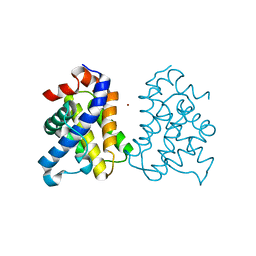 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site.
Mol.Cell, 24, 2006
|
|
2IMS
 
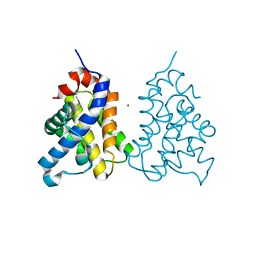 | | The X-ray Structure of a Bak Homodimer Reveals an Inhibitory Zinc Binding Site | | Descriptor: | Apoptosis regulator BAK, ZINC ION | | Authors: | Moldoveanu, T, Liu, Q, Tocilj, A, Watson, M, Shore, G.C, Gehring, K.B. | | Deposit date: | 2006-10-04 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The X-Ray Structure of a BAK Homodimer Reveals an Inhibitory Zinc Binding Site
Mol.Cell, 24, 2006
|
|
5VF6
 
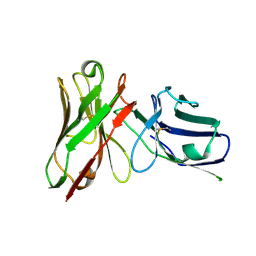 | |
1YQC
 
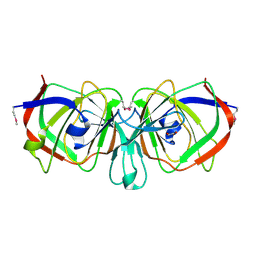 | | Crystal Structure of Ureidoglycolate Hydrolase (AllA) from Escherichia coli O157:H7 | | Descriptor: | GLYOXYLIC ACID, Ureidoglycolate hydrolase | | Authors: | Raymond, S, Tocilj, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Crystal structure of ureidoglycolate hydrolase (AllA) from Escherichia coli O157:H7
Proteins, 61, 2005
|
|
1FKA
 
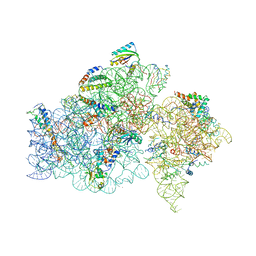 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
1JZY
 
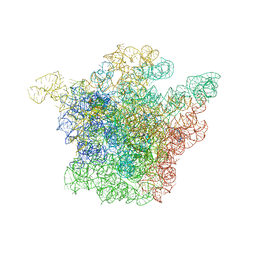 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, ERYTHROMYCIN A, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1K01
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, CHLORAMPHENICOL, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1JZZ
 
 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | Descriptor: | 23S rRNA, MAGNESIUM ION, ROXITHROMYCIN, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2001-09-17 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1J5A
 
 | | STRUCTURAL BASIS FOR THE INTERACTION OF ANTIBIOTICS WITH THE PEPTIDYL TRANSFERASE CENTER IN EUBACTERIA | | Descriptor: | 23S RRNA, CLARITHROMYCIN, MAGNESIUM ION, ... | | Authors: | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | Deposit date: | 2002-03-06 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
