1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
2MOM
 
 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
2MOF
 
 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
3GAT
 
 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
5T1N
 
 | |
1B4C
 
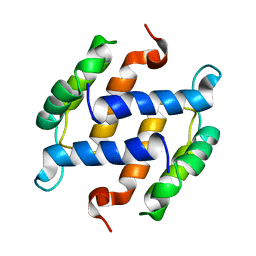 | | SOLUTION STRUCTURE OF RAT APO-S100B USING DIPOLAR COUPLINGS | | Descriptor: | PROTEIN (S-100 PROTEIN, BETA CHAIN) | | Authors: | Weber, D.J, Drohat, A.C, Tjandra, N, Baldisseri, D.M. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The use of dipolar couplings for determining the solution structure of rat apo-S100B(betabeta).
Protein Sci., 8, 1999
|
|
1KHM
 
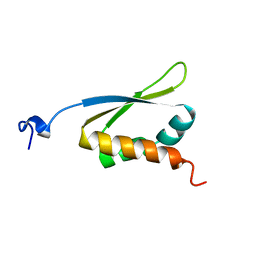 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
1CMZ
 
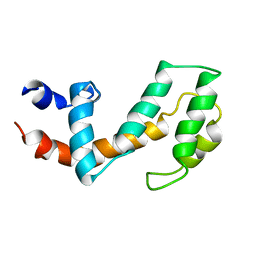 | |
1SN6
 
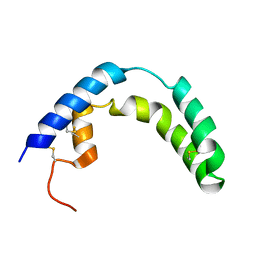 | |
1NAJ
 
 | | High resolution NMR Structure Of DNA Dodecamer Determined In Aqueous Dilute Liquid Crystalline Phase | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Wu, Z, Delaglio, F, Tjandra, N, Zhurkin, V, Bax, A. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and (31)P chemical shift anisotropy.
J.Biomol.Nmr, 26, 2003
|
|
1PC2
 
 | |
5T1O
 
 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
5VKG
 
 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with tenatoprazole | | Descriptor: | 4-methoxy-1-(5-methoxy-3H-imidazo[4,5-b]pyridin-2-yl)-3,5-dimethyl-2-(sulfanylmethyl)pyridin-1-ium, Tumor susceptibility gene 101 protein | | Authors: | Strickland, M, Ehrlich, L.S, Watanabe, S, Khan, M, Strub, M.-P, Luan, C.H, Powell, M.D, Leis, J, Tjandra, N, Carter, C. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Tsg101 chaperone function revealed by HIV-1 assembly inhibitors.
Nat Commun, 8, 2017
|
|
1SNL
 
 | |
1SSE
 
 | |
1F16
 
 | |
6CMY
 
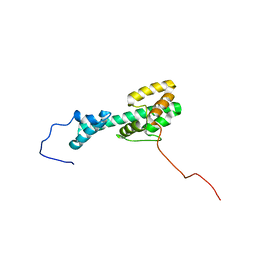 | | Solution NMR Structure Determination of Mouse Melanoregulin | | Descriptor: | Melanoregulin | | Authors: | Rout, A.K, Wu, X, Strub, M.P, Starich, M.R, Hammer III, J.A, Tjandra, N. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of Melanoregulin Reveals a Role for Cholesterol Recognition in the Protein's Ability to Promote Dynein Function.
Structure, 26, 2018
|
|
1CFD
 
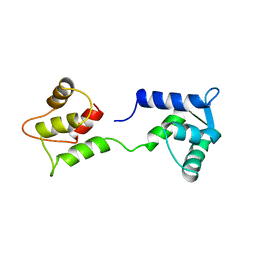 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-10-18 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1CFC
 
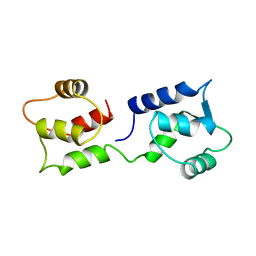 | | CALCIUM-FREE CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Kuboniwa, H, Tjandra, N, Grzesiek, S, Ren, H, Klee, C.B, Bax, A. | | Deposit date: | 1995-08-02 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-free calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
6BYV
 
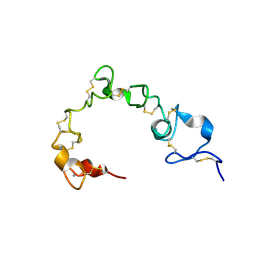 | | Solution NMR structure of cysteine-rich calcium bound domains of very low density lipoprotein receptor | | Descriptor: | CALCIUM ION, Very low-density lipoprotein receptor | | Authors: | Banerjee, K, Gruschus, J.M, Tjandra, N, Yakovlev, S, Medved, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Recombinant Fragment Containing Three Fibrin-Binding Cysteine-Rich Domains of the Very Low Density Lipoprotein Receptor.
Biochemistry, 57, 2018
|
|
6UD0
 
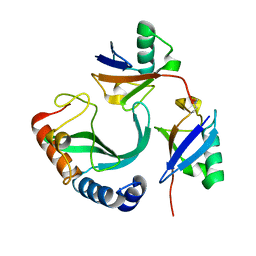 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
1MFS
 
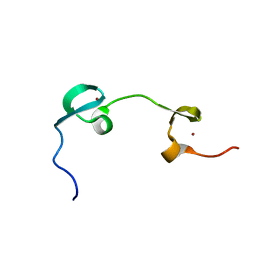 | | DYNAMICAL BEHAVIOR OF THE HIV-1 NUCLEOCAPSID PROTEIN; NMR, 30 STRUCTURES | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Turner, B.G, De Guzman, R.N, Lee, B.M, Tjandra, N. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dynamical behavior of the HIV-1 nucleocapsid protein.
J.Mol.Biol., 279, 1998
|
|
1M12
 
 | |
1G03
 
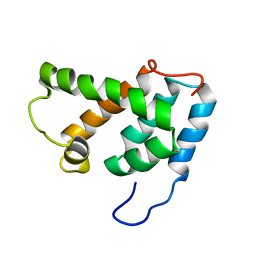 | | NMR STRUCTURE OF N-TERMINAL DOMAIN OF HTLV-I CA1-134 | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Cornilescu, C.C, Bouamr, F, Yao, X, Carter, C, Tjandra, N. | | Deposit date: | 2000-10-05 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein.
J.Mol.Biol., 306, 2001
|
|
8V3N
 
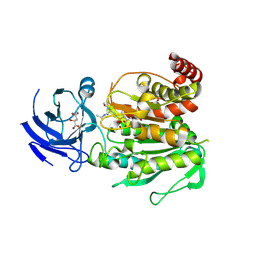 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
