6H65
 
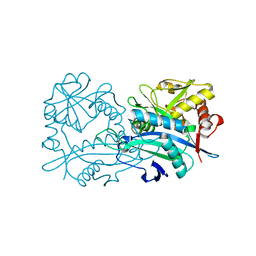 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
7YWS
 
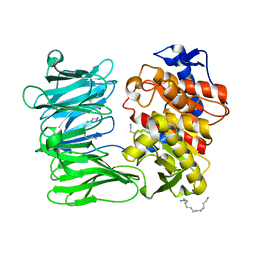 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 3 spermine molecules at 1.7 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
7YX7
 
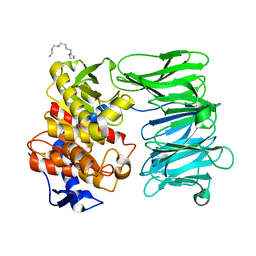 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 1 spermine molecule at 1.72 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
7YWZ
 
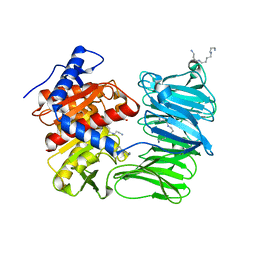 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution | | Descriptor: | GLYCEROL, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution
To Be Published
|
|
7YWP
 
 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
7ZJZ
 
 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-04-12 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
2NDP
 
 | | Structure of DNA-binding HU protein from micoplasma Mycoplasma gallisepticum | | Descriptor: | Histone-like DNA-binding superfamily protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I, Popov, V.O. | | Deposit date: | 2016-09-13 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enhanced conformational flexibility of the histone-like (HU) protein from Mycoplasma gallisepticum.
J.Biomol.Struct.Dyn., 36, 2018
|
|
7A68
 
 | | proteinase K crystallized from 0.5 M NaNO3 | | Descriptor: | Proteinase K, SODIUM ION | | Authors: | Ilina, K.B, Kulikov, A.G, Timofeev, V.I, Marchenkova, M.A, Pisarevsky, Y.V, Kovalchuk, M.V. | | Deposit date: | 2020-08-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | proteinase K crystallized from 0.5 M NaNO3
To Be Published
|
|
7AKN
 
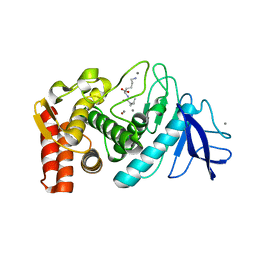 | | Thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Boikova, A.S, Ilina, K.B, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2020-10-01 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Thermolysin from Bacillus thermoproteolyticus
To Be Published
|
|
4DUK
 
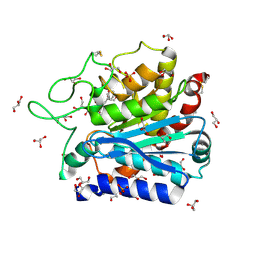 | | Carboxypeptidase T with L-BENZYLSUCCINIC ACID | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-02-22 | | Release date: | 2013-02-27 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural insights into the broad substrate specificity of carboxypeptidase T from Thermoactinomyces vulgaris.
Febs J., 282, 2015
|
|
4DJL
 
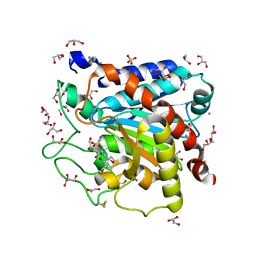 | | Carboxypeptidase T with N-sulfamoyl-L-phenylalanine | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-02-02 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Carboxypeptidase T with N-sulfamoyl-L-phenylalanine
TO BE PUBLISHED
|
|
6QWX
 
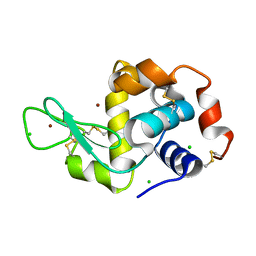 | | HEWL lysozyme, crystallized from NiCl2 solution | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWW
 
 | | HEWL lysozyme, crystallized from CuCl2 solution | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWY
 
 | | HEWL lysozyme, crystallized from NaCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWZ
 
 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QX0
 
 | | HEWL lysozyme, crystallized from LiCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
2HN9
 
 | |
