8B0Q
 
 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
2V1C
 
 | |
2VF8
 
 | | Crystal structure of UvrA2 from Deinococcus radiodurans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EXCINUCLEASE ABC SUBUNIT A, PHOSPHATE ION, ... | | Authors: | Timmins, J, Gordon, E, Caria, S, Leonard, G, Kuo, M.S, Monchois, V, McSweeney, S. | | Deposit date: | 2007-10-31 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mutational Analyses of Deinococcus Radiodurans Uvra2 Provide Insight Into DNA Binding and Damage Recognition by Uvras.
Structure, 17, 2009
|
|
2VF7
 
 | | Crystal structure of UvrA2 from Deinococcus radiodurans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EXCINUCLEASE ABC, SUBUNIT A., ... | | Authors: | Timmins, J, Gordon, E, Caria, S, Leonard, G, Kuo, M.S, Monchois, V, McSweeney, S. | | Deposit date: | 2007-10-31 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs.
Structure, 17, 2009
|
|
2BHU
 
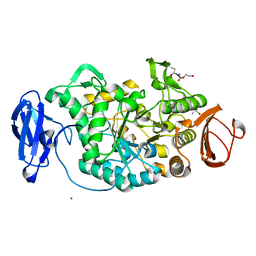 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MALTOOLIGOSYLTREHALOSE TREHALOHYDROLASE, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-18 | | Release date: | 2005-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BHZ
 
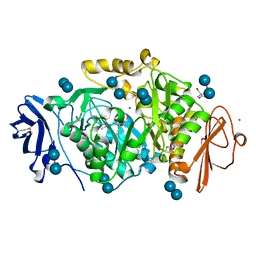 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with maltose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
2BHY
 
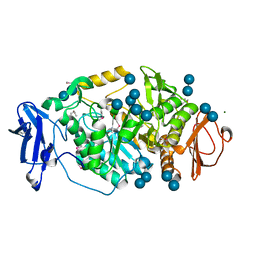 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with trehalose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
7QVB
 
 | |
4JCV
 
 | |
4ZC0
 
 | | Structure of a dodecameric bacterial helicase | | Descriptor: | HEXATANTALUM DODECABROMIDE, Replicative DNA helicase | | Authors: | Bazin, A, Cherrier, M.V, Gutsche, I, Timmins, J, Terradot, L. | | Deposit date: | 2015-04-15 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structure and primase-mediated activation of a bacterial dodecameric replicative helicase.
Nucleic Acids Res., 43, 2015
|
|
4UOB
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
4UNF
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-1 | | Descriptor: | ACETATE ION, ENDONUCLEASE III-1, IRON/SULFUR CLUSTER, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
1W3S
 
 | | The crystal structure of RecO from Deinococcus radiodurans. | | Descriptor: | HYPOTHETICAL PROTEIN DR0819, ZINC ION | | Authors: | Leiros, I, Timmins, J, Hall, D.R, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-18 | | Release date: | 2005-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and DNA-Binding Analysis of Reco from Deinococcus Radiodurans
Embo J., 24, 2005
|
|
1T3E
 
 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
2YG8
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA-3-methyladenine glycosidase II, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
2YG9
 
 | | Structure of an unusual 3-Methyladenine DNA Glycosylase II (Alka) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, DNA-3-methyladenine glycosidase II, putative, ... | | Authors: | Moe, E, Hall, D.R, Leiros, I, Talstad, V, Timmins, J, McSweeney, S. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
4AD8
 
 | | Crystal structure of a deletion mutant of Deinococcus radiodurans RecN | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABX
 
 | | Crystal structure of Deinococcus radiodurans RecN coiled-coil domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABY
 
 | | Crystal structure of Deinococcus radiodurans RecN head domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4C30
 
 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, form 2 | | Descriptor: | DNA HELICASE II, DNA STRAND FOR25, DNA STRAND REV25, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4C2T
 
 | | Crystal structure of full length Deinococcus radiodurans UvrD in complex with DNA | | Descriptor: | DNA HELICASE II, DNA STRAND FOR28, DNA STRAND REV28, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.997 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4C2U
 
 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, Form 1 | | Descriptor: | DNA HELICASE II, FOR25, GLYCEROL, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4A1F
 
 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | Descriptor: | CITRATE ANION, REPLICATIVE DNA HELICASE | | Authors: | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
2BY2
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY1
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-28 | | Release date: | 2006-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
