4AQR
 
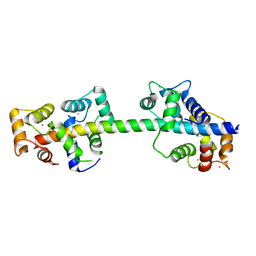 | | Crystal structure of calmodulin in complex with the regulatory domain of a plasma-membrane Ca2+-ATPase | | Descriptor: | CALCIUM ION, CALCIUM-TRANSPORTING ATPASE 8, PLASMA MEMBRANE-TYPE, ... | | Authors: | Tidow, H, Poulsen, L.R, Andreeva, A, Hein, K.L, Palmgren, M.G, Nissen, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Bimodular Mechanism of Calcium Control in Eukaryotes
Nature, 491, 2012
|
|
1UUC
 
 | |
2UWQ
 
 | |
2VY4
 
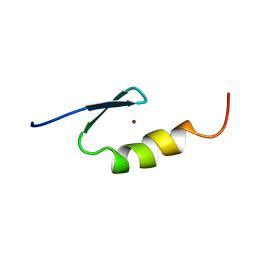 | | U11-48K CHHC ZN-FINGER DOMAIN | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-17 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
2VY5
 
 | | U11-48K CHHC Zn-finger protein domain | | Descriptor: | U11/U12 SMALL NUCLEAR RIBONUCLEOPROTEIN 48 KDA PROTEIN, ZINC ION | | Authors: | Tidow, H, Andreeva, A, Rutherford, T.J, Fersht, A.R. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns.
Structure, 17, 2009
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2FEJ
 
 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2NNT
 
 | | General structural motifs of amyloid protofilaments | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Ferguson, N, Becker, J, Tidow, H, Tremmel, S, Sharpe, T.D, Krause, G, Flinders, J, Petrovich, M, Berriman, J, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | General structural motifs of amyloid protofilaments.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5EDL
 
 | | Crystal structure of an S-component of ECF transporter | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Putative HMP/thiamine permease protein YkoE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2015-10-21 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a Group I Energy Coupling Factor Vitamin Transporter S Component in Complex with Its Cognate Substrate.
Cell Chem Biol, 23, 2016
|
|
6HCS
 
 | |
6H01
 
 | |
5NOH
 
 | | HRSV M2-1 core domain | | Descriptor: | Transcription elongation factor M2-1 | | Authors: | Josts, I, Almeida Hernandez, Y, Molina, I.G, de Prat-Gay, G, Tidow, H. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5NKX
 
 | | HRSV M2-1 core domain, P3221 crystal form | | Descriptor: | M2-1 | | Authors: | Almeida Hernandez, Y, Josts, I, Molina, I.G, de Pray-Gay, G, Tidow, H. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.00008678 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8AZC
 
 | |
6Z8A
 
 | | Outer membrane FoxA in complex with nocardamine | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Nocardamine-Dependent Iron Uptake in Pseudomonas aeruginosa : Exclusive Involvement of the FoxA Outer Membrane Transporter.
Acs Chem.Biol., 15, 2020
|
|
8B43
 
 | | Crystal structure of ferrioxamine transporter | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Ferrichrome-iron receptor, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6I98
 
 | |
6I96
 
 | |
8AZD
 
 | |
8AZL
 
 | |
8AZN
 
 | |
8AZI
 
 | |
