6QV5
 
 | |
5A67
 
 | |
2HQT
 
 | |
5IIG
 
 | |
4LSA
 
 | | Crystal structure of BRI1 sud1 (Gly643Glu) bound to brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
5IJH
 
 | |
6R1H
 
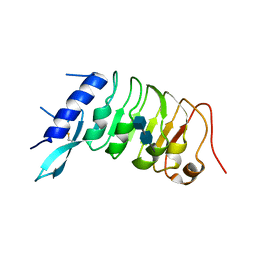 | |
5IIQ
 
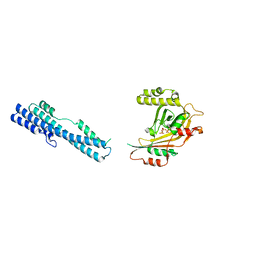 | |
5IJJ
 
 | |
5IJP
 
 | |
2HRA
 
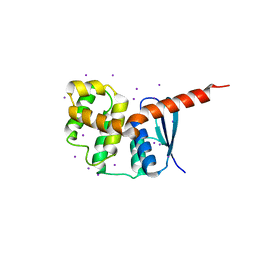 | | Crystal structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA aminoacylation and nuclear export cofactor Arc1p reveal a novel function for an old fold | | Descriptor: | Glutamyl-tRNA synthetase, cytoplasmic, IODIDE ION | | Authors: | Simader, H, Hothorn, M, Suck, D. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
6QVA
 
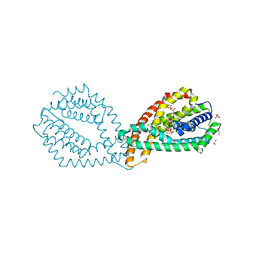 | |
5LNC
 
 | |
4LSC
 
 | | Isolated SERK1 co-receptor ectodomain at high resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Somatic embryogenesis receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
1FT9
 
 | | STRUCTURE OF THE REDUCED (FEII) CO-SENSING PROTEIN FROM R. RUBRUM | | Descriptor: | CARBON MONOXIDE OXIDATION SYSTEM TRANSCRIPTION REGULATOR, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanzilotta, W.N, Schuller, D.J, Poulos, T.L, Thorsteinsson, M.V, Kirby, B, Roberts, G.P. | | Deposit date: | 2000-09-12 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the CO sensing transcription activator CooA.
Nat.Struct.Biol., 7, 2000
|
|
3DXB
 
 | | Structure of the UHM domain of Puf60 fused to thioredoxin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, thioredoxin N-terminally fused to Puf60(UHM) | | Authors: | Corsini, L, Hothorn, M, Scheffzek, K, Stier, G, Sattler, M. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimerization and Protein Binding Specificity of the U2AF Homology Motif of the Splicing Factor Puf60.
J.Biol.Chem., 284, 2009
|
|
3G3T
 
 | | Crystal structure of a eukaryotic polyphosphate polymerase in complex with orthophosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Vacuolar transporter chaperone 4 | | Authors: | Lenherr, E.D, Hothorn, M, Scheffzek, K. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase.
Science, 324, 2009
|
|
2PE8
 
 | |
2PEH
 
 | | Crystal structure of the UHM domain of human SPF45 in complex with SF3b155-ULM5 | | Descriptor: | Splicing factor 3B subunit 1, Splicing factor 45 | | Authors: | Corsini, L, Basquin, J, Hothorn, M, Sattler, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | U2AF-homology motif interactions are required for alternative splicing regulation by SPF45.
Nat.Struct.Mol.Biol., 14, 2007
|
|
