1NS2
 
 | |
1NSS
 
 | |
1NS0
 
 | |
1NS7
 
 | |
1P90
 
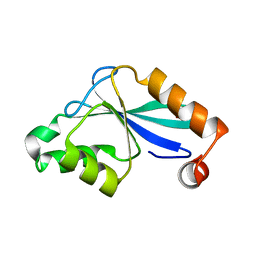 | | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 resolution | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein | | Authors: | Dyer, D.H, Rubio, L.M, Thoden, J.B, Holden, H.M, Ludden, P.W, Rayment, I. | | Deposit date: | 2003-05-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-dimensional Structure of the Core Domain of NafY from Azotobacter vinelandii determined at 1.8 A resolution
J.Biol.Chem., 278, 2003
|
|
3FSB
 
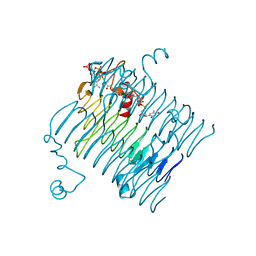 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with CoA and dTDP-3-amino-quinovose | | Descriptor: | COENZYME A, QdtC, [(3R,4S,5S,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
3FSC
 
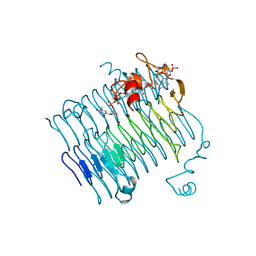 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with CoA and dTDP-3-amino-fucose | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, COENZYME A, QdtC | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
3FS8
 
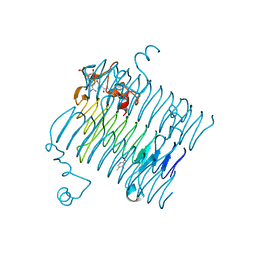 | | Crystal structure of QdtC, the dTDP-3-amino-3,6-dideoxy-D-glucose N-acetyl transferase from Thermoanaerobacterium thermosaccharolyticum in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, QdtC, THYMINE | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of QdtC: an N-Acetyltransferase Required for the Biosynthesis of dTDP-3-Acetamido-3,6-Dideoxy-alpha-D-Glucose.
Biochemistry, 48, 2009
|
|
7JID
 
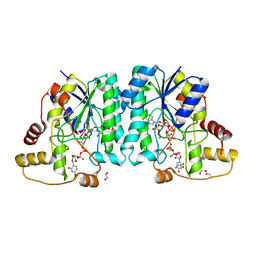 | | Crystal structure of the L780 UDP-rhamnose synthase from Acanthamoeba polyphaga mimivirus | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-L-Rhamnose Synthase, ... | | Authors: | Bockhaus, N.J, Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The high-resolution structure of a UDP-L-rhamnose synthase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
7L7Y
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of UDP and acetyl-conezyme A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L82
 
 | | x-ray structure of the psychrobacter cryohalolentis Pcryo_0637 N-acetyltransferase in the presene of its reaction tetrahedral intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Putative acetyl transferase protein, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L81
 
 | | x-ray structure of the psychrobacter cryohalolentis N-acetyltransferase Pcryo_0637 in the presence of coenzyme A and | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7X
 
 | | X-ray structure of the Pcryo_0638 aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7Z
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of coenzyme A and UDP-di-N-acetyl-bacillosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
1MUS
 
 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
