6K31
 
 | |
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
1ULJ
 
 | | Biphenyl dioxygenase (BphA1A2) in complex with the substrate | | Descriptor: | BIPHENYL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Furusawa, Y, Nagarajan, V, Masai, E, Tanokura, M, Fukuda, M, Senda, T. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Terminal Oxygenase Component of Biphenyl Dioxygenase Derived from Rhodococcus sp. Strain RHA1
J.Mol.Biol., 342, 2004
|
|
1IJ5
 
 | | METAL-FREE STRUCTURE OF MULTIDOMAIN EF-HAND PROTEIN, CBP40, FROM TRUE SLIME MOLD | | Descriptor: | PLASMODIAL SPECIFIC LAV1-2 PROTEIN | | Authors: | Iwasaki, W, Sasaki, H, Nakamura, A, Kohama, K, Tanokura, M. | | Deposit date: | 2001-04-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Metal-Free and Ca(2+)-Bound Structures of a Multidomain EF-Hand Protein, CBP40, from the Lower Eukaryote Physarum polycephalum
Structure, 11, 2003
|
|
1X25
 
 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
1ZR7
 
 | |
1Y49
 
 | |
7WAB
 
 | | Crystal structure of the prolyl endoprotease, PEP, from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPASS (Complex proteins associated with Set1p) component shg1 family protein, ... | | Authors: | Miyazono, K, Kubota, K, Takahashi, K, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate recognition mechanism of the prolyl endoprotease PEP from Aspergillus niger.
Biochem.Biophys.Res.Commun., 591, 2022
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
5Z7Y
 
 | | Crystal structure of Striga hermonthica HTL7 (ShHTL7) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Hyposensitive to light 7, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7Z
 
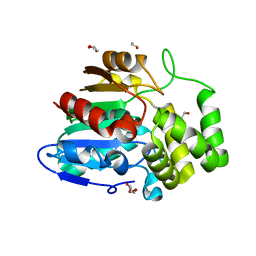 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5ZD4
 
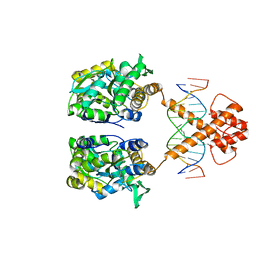 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | Authors: | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | Deposit date: | 2018-02-22 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
3OIT
 
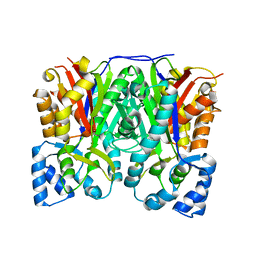 | | Crystal structure of curcuminoid synthase CUS from Oryza sativa | | Descriptor: | Os07g0271500 protein | | Authors: | Miyazono, K, Um, J, Imai, F.L, Katsuyama, Y, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of curcuminoid synthase CUS from Oryza sativa
Proteins, 79, 2011
|
|
3ORY
 
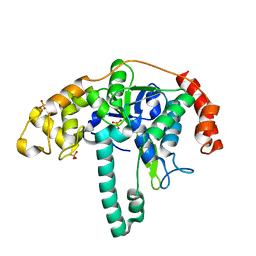 | | Crystal structure of Flap endonuclease 1 from hyperthermophilic archaeon Desulfurococcus amylolyticus | | Descriptor: | PHOSPHATE ION, flap endonuclease 1 | | Authors: | Mase, T, Kubota, K, Miyazono, K, Kawarabayashii, Y, Tanokura, M. | | Deposit date: | 2010-09-08 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1V5I
 
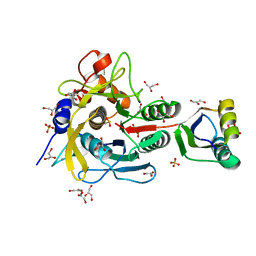 | | Crystal structure of serine protease inhibitor POIA1 in complex with subtilisin BPN' | | Descriptor: | CALCIUM ION, GLYCEROL, IA-1=serine proteinase inhibitor, ... | | Authors: | Lee, W.C, Kikkawa, M, Kojima, S, Miura, K, Tanokura, M. | | Deposit date: | 2003-11-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of serine protease inhibitor POIA1 in complex with subtilisin BPN'
To be Published
|
|
1V3Y
 
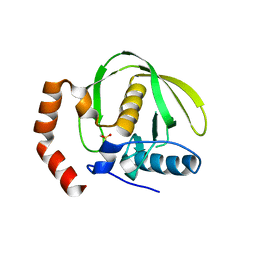 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | Descriptor: | Peptide deformylase | | Authors: | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
2EGD
 
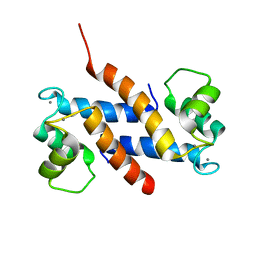 | | Crystal structure of human S100A13 in the Ca2+-bound state | | Descriptor: | CALCIUM ION, Protein S100-A13 | | Authors: | Imai, F.L, Nagata, K, Yonezawa, N, Nakano, M, Tanokura, M. | | Deposit date: | 2007-02-28 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human S100A13 in the Ca2+-bound state
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1DET
 
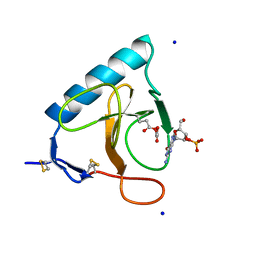 | | RIBONUCLEASE T1 CARBOXYMETHYLATED AT GLU 58 IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Ishikawa, K, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 carboxymethylated at Glu58 in complex with 2'-GMP.
Biochemistry, 35, 1996
|
|
4XRE
 
 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
4YOW
 
 | |
4YOR
 
 | |
7CFA
 
 | |
7CO1
 
 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
4YOV
 
 | |
4HE7
 
 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
