1XF2
 
 | | Structure of Fab DNA-1 complexed with dT3 | | 分子名称: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | 著者 | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | 登録日 | 2004-09-13 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
4U8I
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8P
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A complexed with UDP | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8L
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8O
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant N207A complexed with UDP | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8N
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant F66A complexed with UDP | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8M
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y317A | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | 分子名称: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
2A3P
 
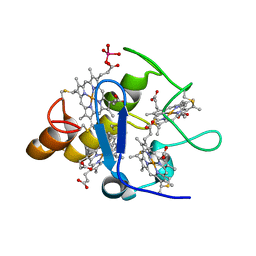 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome with bound molybdate | | 分子名称: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C, ... | | 著者 | Pattarkine, M.V, Lee, Y.-H, Tanner, J.J, Wall, J.D. | | 登録日 | 2005-06-25 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
2A3M
 
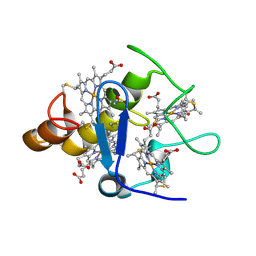 | | Structure of Desulfovibrio desulfuricans G20 tetraheme cytochrome (oxidized form) | | 分子名称: | COG3005: Nitrate/TMAO reductases, membrane-bound tetraheme cytochrome c subunit, HEME C | | 著者 | Pattarkine, M.V, Tanner, J.J, Bottoms, C.A, Lee, Y.H, Wall, J.D. | | 登録日 | 2005-06-25 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Desulfovibrio desulfuricans G20 Tetraheme Cytochrome Structure at 1.5A and Cytochrome Interaction with Metal Complexes
J.Mol.Biol., 358, 2006
|
|
2AY0
 
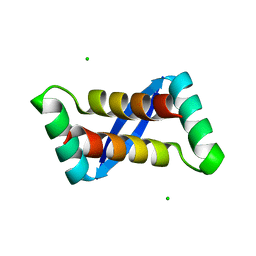 | | Structure of the Lys9Met mutant of the E. coli Proline Utilization A (PutA) DNA-binding domain. | | 分子名称: | Bifunctional putA protein, CHLORIDE ION | | 著者 | Larson, J.D, Schuermann, J.P, Zhou, Y, Jenkins, J.L, Becker, D.F, Tanner, J.J. | | 登録日 | 2005-09-06 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition.
Protein Sci., 15, 2006
|
|
1U8F
 
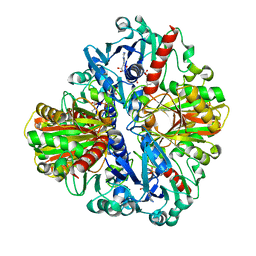 | |
1RWY
 
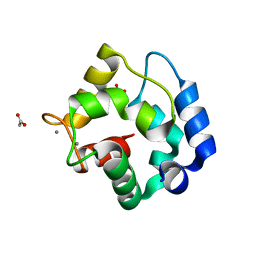 | | CRYSTAL STRUCTURE OF RAT ALPHA-PARVALBUMIN AT 1.05 RESOLUTION | | 分子名称: | ACETIC ACID, AMMONIUM ION, CALCIUM ION, ... | | 著者 | Bottoms, C.A, Schuermann, J.P, Agah, S, Henzl, M.T, Tanner, J.J. | | 登録日 | 2003-12-17 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Structure of Rat Alpha-Parvalbumin at 1.05 Resolution
Protein Sci., 13, 2004
|
|
4DSG
 
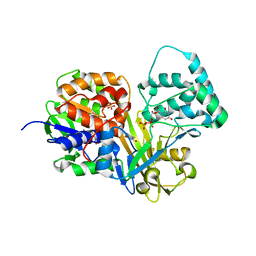 | | Crystal Structure of oxidized UDP-Galactopyranose mutase | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | 著者 | Singh, H, Dhatwalia, R, Tanner, J.J. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.249 Å) | | 主引用文献 | Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Biochemistry, 51, 2012
|
|
4DSH
 
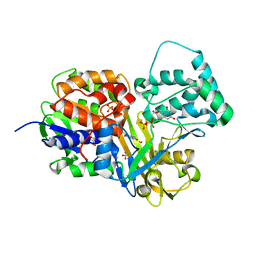 | | Crystal structure of reduced UDP-Galactopyranose mutase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Dhatwalia, R, Singh, H, Tanner, J.J. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structures of Trypanosoma cruzi UDP-Galactopyranose Mutase Implicate Flexibility of the Histidine Loop in Enzyme Activation.
Biochemistry, 51, 2012
|
|
5T19
 
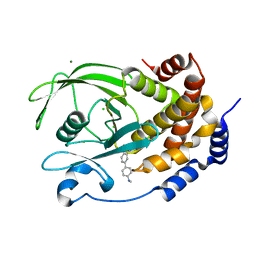 | | Structure of PTP1B complexed with N-(3'-(1,1-dioxido-4-oxo-1,2,5-thiadiazolidin-2-yl)-4'-methyl-[1,1'-biphenyl]-4-yl)acetamide | | 分子名称: | 5-[4-methyl-4'-(methylamino)[1,1'-biphenyl]-3-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Laciak, A.R, Tanner, J.J. | | 登録日 | 2016-08-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1001 Å) | | 主引用文献 | Covalent Allosteric Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) by an Inhibitor-Electrophile Conjugate.
Biochemistry, 56, 2017
|
|
4LH2
 
 | |
4LH0
 
 | |
4LH1
 
 | |
4LH3
 
 | |
4LGZ
 
 | |
4NM9
 
 | |
4NMA
 
 | |
4NMD
 
 | |
