1UI7
 
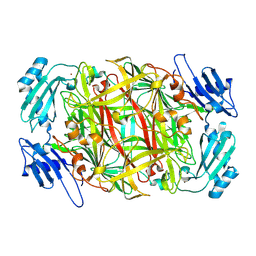 | | Site-directed mutagenesis of His433 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1UI8
 
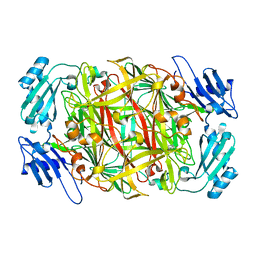 | | Site-directed mutagenesis of His592 involved in binding of copper ion in Arthrobacter globiformis amine oxidase | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Matsunami, H, Okajima, T, Hirota, S, Yamaguchi, H, Hori, H, Kuroda, S, Tanizawa, K. | | Deposit date: | 2003-07-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical rescue of a site-specific mutant of bacterial copper amine oxidase for generation of the topa quinone cofactor
Biochemistry, 43, 2004
|
|
1WXR
 
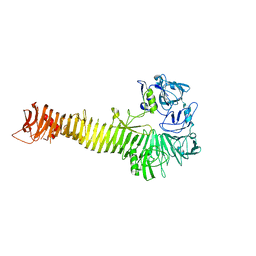 | | Crystal structure of Heme Binding protein, an autotransporter hemoglobine protease from pathogenic Escherichia coli | | Descriptor: | haemoglobin protease | | Authors: | Otto, B.R, Sijbrandi, R, Luirink, J, Oudega, B, Heddle, J.G, Mizutani, K, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic escherichia coli
J.Biol.Chem., 280, 2005
|
|
1VD6
 
 | |
2ZFH
 
 | |
1WN1
 
 | | Crystal Structure of Dipeptiase from Pyrococcus Horikoshii OT3 | | Descriptor: | COBALT (II) ION, dipeptidase | | Authors: | Jeyakanthan, J, Taka, J, Kitaguchi, Y, Shiro, Y, Yokoyama, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-26 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dipeptiase from Pyrococcus Horikoshii OT3
To be Published
|
|
1V9K
 
 | | The crystal structure of the catalytic domain of pseudouridine synthase RluC from Escherichia coli | | Descriptor: | Ribosomal large subunit pseudouridine synthase C, SULFATE ION | | Authors: | Machida, Y, Mizutani, K, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1V4P
 
 | |
1VC2
 
 | |
2ZQ9
 
 | | Cephalothin acyl-intermediate structure of class a beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase Toho-1, CEPHALOTHIN, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
1JJU
 
 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2E2V
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with benzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2CWU
 
 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2ELG
 
 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2E2T
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with phenylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E2U
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with 4-hydroxybenzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2D1W
 
 | | Substrate Schiff-Base intermediate with tyramine in copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Kuroda, S, Nakamoto, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2005-09-01 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction
Biochem.Biophys.Res.Commun., 342, 2006
|
|
2CWV
 
 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2E66
 
 | | Crystal Structure Of CutA1 From Pyrococcus Horikoshii OT3, Mutation D60A | | Descriptor: | CHLORIDE ION, Divalent-cation tolerance protein cutA, SODIUM ION | | Authors: | Bagautdinov, B, Sawano, M, Bagautdinova, S, Yutani, K, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the hyper-thermostability of CutA1
To be Published
|
|
2HOW
 
 | |
1G3B
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE MAGNESIUM(II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3D
 
 | | BOVINE BETA-TRYPSIN BOUND TO META-AMIDINO SCHIFF BASE COPPER (II) CHELATE | | Descriptor: | 2-(5-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
1G3C
 
 | | BOVINE BETA-TRYPSIN BOUND TO PARA-AMIDINO SCHIFF BASE IRON(III) CHELATE | | Descriptor: | 2-(4-CARBAMIMIDOYL-2-HYDROXY-BENZYLAMINO)-PROPIONIC ACID, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Toyota, E, Ng, K.K.S, Sekizaki, H, Itoh, K, Tanizawa, K, James, M.N.G. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analyses of complexes between bovine beta-trypsin and Schiff base copper(II) or iron(III) chelates.
J.Mol.Biol., 305, 2001
|
|
