3TTI
 
 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3B6Z
 
 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN 90% ACETONITRILE-WATER | | Descriptor: | ACETONITRILE, LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-06 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
1JIR
 
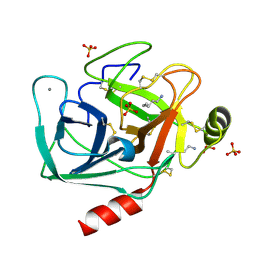 | | Crystal Structure of Trypsin Complex with Amylamine in Cyclohexane | | Descriptor: | AMYLAMINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Wu, G, Zhu, G, Huang, Q, Qian, M, Tang, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of beta-Trypsin Complex with Amylamine in Cyclohexane
To be Published
|
|
1L5J
 
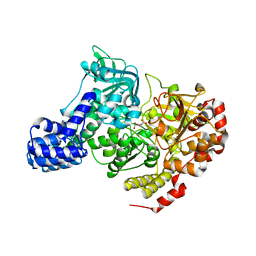 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
4ZBJ
 
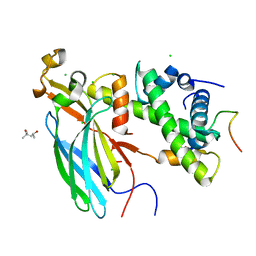 | | UBN1 peptide bound to H3.3/H4/Asf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Marmorstein, R, Ricketts, M.D, Tang, Y. | | Deposit date: | 2015-04-14 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex.
Nat Commun, 6, 2015
|
|
6DF4
 
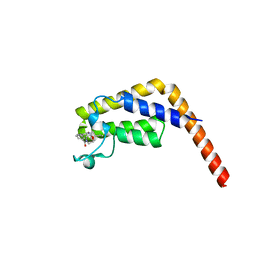 | | TAF1-BD2 in complex with Cpd8 (6-(but-3-en-1-yl)-4-(3-(morpholine-4-carbonyl)phenyl)-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 6-(but-3-en-1-yl)-4-[3-(morpholine-4-carbonyl)phenyl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2018-05-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GNE-371, a Potent and Selective Chemical Probe for the Second Bromodomains of Human Transcription-Initiation-Factor TFIID Subunit 1 and Transcription-Initiation-Factor TFIID Subunit 1-like.
J. Med. Chem., 61, 2018
|
|
4LYO
 
 | | CROSS-LINKED CHICKEN LYSOZYME CRYSTAL IN NEAT ACETONITRILE, THEN BACK-SOAKED IN WATER | | Descriptor: | LYSOZYME | | Authors: | Huang, Q, Wang, Z, Zhu, G, Qian, M, Shao, M, Jia, Y, Tang, Y. | | Deposit date: | 1998-03-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray studies on cross-linked lysozyme crystals in acetonitrile-water mixture.
Biochim.Biophys.Acta, 1384, 1998
|
|
4RO5
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM | | Descriptor: | GLYCEROL, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-27 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4RPM
 
 | | Crystal structure of the SAT domain from the non-reducing fungal polyketide synthase CazM with bound hexanoyl | | Descriptor: | HEXANOIC ACID, HEXANOYL-COENZYME A, SAT domain from CazM | | Authors: | Winter, J.M, Cascio, D, Sawaya, M.R, Tang, Y. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Basis for Controlling Chemical Modularity in Fungal Polyketide Biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
4K2X
 
 | |
4LCM
 
 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD9 mutant (simh9014) | | Descriptor: | Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4LCL
 
 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD6 mutant (simh6208) | | Descriptor: | ISOPROPYL ALCOHOL, Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
3TFZ
 
 | | Crystal structure of Zhui aromatase/cyclase from Streptomcyes sp. R1128 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclase, POTASSIUM ION | | Authors: | Ames, B.D, Lee, M.Y, Moody, C, Zhang, W, Tang, Y, Wong, S.K, Tsai, S.C. | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Characterization of ZhuI Aromatase/Cyclase from the R1128 Polyketide Pathway.
Biochemistry, 50, 2011
|
|
3TKK
 
 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
3SKV
 
 | | Salicylyl-Acyltransferase SsfX3 from a Tetracycline Biosynthetic Pathway | | Descriptor: | GLYCEROL, SsfX3 | | Authors: | Pickens, L.B, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2011-06-23 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Biochemical Characterization of the Salicylyl-acyltranferase SsfX3 from a Tetracycline Biosynthetic Pathway.
J.Biol.Chem., 286, 2011
|
|
2I32
 
 | |
2LFC
 
 | | Solution NMR Structure of Fumarate reductase flavoprotein subunit from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR145J | | Descriptor: | Fumarate reductase, flavoprotein subunit | | Authors: | Liu, G, Tang, Y, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Acton, T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target LpR145J
To be Published
|
|
6BQD
 
 | | TAF1-BD2 bromodomain in complex with (E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
6BQA
 
 | | BRD9 bromodomain in complex with 3-(6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.031 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
7BQJ
 
 | | The structure of PdxI | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyltransf_2 domain-containing protein, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQO
 
 | | The structure of HpiI in complex with its substrate analogue | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQP
 
 | | The structure of HpiI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-25 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
7BQK
 
 | | The structure of PdxI in complex with its substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 3-[(E,2S,4S)-2,4-dimethyloct-6-enoyl]-4-oxidanyl-1H-pyridin-2-one, GLYCEROL, ... | | Authors: | Cai, Y.J, Ohashi, M, Zhou, J.H, Tang, Y. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An enzymatic Alder-ene reaction.
Nature, 586, 2020
|
|
