3RXU
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and A06, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXI
 
 | | Crystal structure of Trypsin complexed with 2-(1H-indol-3-yl)ethanamine | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXL
 
 | | Crystal structure of Trypsin complexed with (2,5-dimethyl-3-furyl)methanamine | | Descriptor: | 1-(2,5-dimethylfuran-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXV
 
 | | Crystal structure of Trypsin complexed with benzamide (F05 and F03, cocktail experiment) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXE
 
 | | Crystal structure of Trypsin complexed with benzamide | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXS
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine (F04 and A06, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
5B4A
 
 | | Crystal structure of LpxH with lipid X in spacegroup P21 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
5B4B
 
 | | Crystal structure of LpxH with lipid X in spacegroup C2 | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
1GD6
 
 | | STRUCTURE OF THE BOMBYX MORI LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Matsuura, A, Aizawa, T, Yao, M, Kawano, K, Tanaka, I, Nitta, K. | | Deposit date: | 2000-09-19 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of an insect lysozyme exhibiting catalytic efficiency at low temperatures.
Biochemistry, 41, 2002
|
|
5B4D
 
 | | Crystal structure of H10N mutant of LpxH | | Descriptor: | GLYCEROL, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
1ITW
 
 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-02-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1TAB
 
 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
2DGD
 
 | | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii | | Descriptor: | 223aa long hypothetical arylmalonate decarboxylase, GLYCEROL | | Authors: | Tanaka, Y, Sasaki, T, Tanabe, E, Yao, M, Tanaka, I, Kumagai, I, Tsumoto, K. | | Deposit date: | 2006-03-10 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of ST0656, a function unknown protein from Sulfolobus tokodaii
To be Published
|
|
2DSO
 
 | | Crystal structure of D138N mutant of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Drp35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-07-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG0
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus | | Descriptor: | DrP35 | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2DG1
 
 | | Crystal structure of Drp35, a 35kDa drug responsive protein from Staphylococcus aureus, complexed with Ca2+ | | Descriptor: | CALCIUM ION, DrP35, GLYCEROL | | Authors: | Tanaka, Y, Ohki, Y, Morikawa, K, Yao, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and Mutational Analyses of Drp35 from Staphylococcus aureus: A POSSIBLE MECHANISM FOR ITS LACTONASE ACTIVITY
J.Biol.Chem., 282, 2007
|
|
2E7D
 
 | | Crystal structure of a NEAT domain from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, Hypothetical protein IsdH, ... | | Authors: | Suenaga, A, Tanaka, Y, Yao, M, Kumagai, I, Tanaka, I, Tsumoto, K. | | Deposit date: | 2007-01-09 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus
J.Biol.Chem., 283, 2008
|
|
1WQ2
 
 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3ANZ
 
 | | Crystal Structure of alpha-hemolysin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Alpha-hemolysin | | Authors: | Yamashita, K, Kawauchi, H, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2010-09-16 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal alpha-hemolysin into heptameric pore structure
Protein Sci., 20, 2011
|
|
2D6Y
 
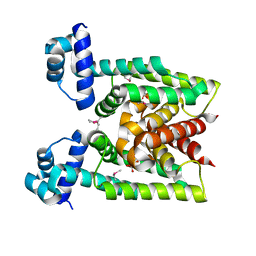 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
2DG7
 
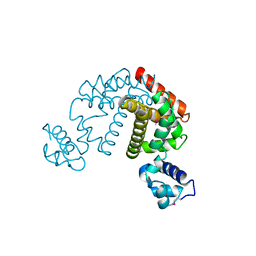 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2DG6
 
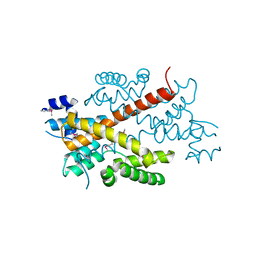 | | Crystal structure of the putative transcriptional regulator SCO5550 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape
Biochem. Biophys. Res. Commun., 435, 2013
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
