6MWV
 
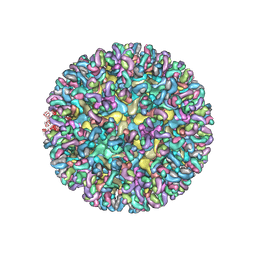 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-58 Antibody | | Descriptor: | E1, E2, EEEV-58 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
1MRJ
 
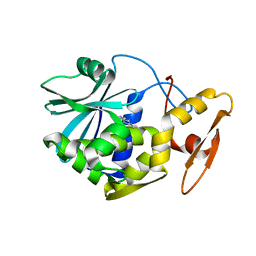 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
6MX4
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
5HJ7
 
 | | Glutamate Racemase Mycobacterium tuberculosis (MurI) with bound D-glutamate, 2.3 Angstrom resolution, X-ray diffraction | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Poen, S, Nakatani, Y, Krause, K. | | Deposit date: | 2016-01-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
7DO6
 
 | |
2RVP
 
 | | Solution structure of DNA Containing Metallo-Base-Pair | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*AP*CP*TP*TP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*AP*AP*CP*TP*AP*TP*AP*TP*TP*A)-3'), SILVER ION | | Authors: | Dairaku, T, Furuita, K, Sato, H, Sebera, J, Nakashima, K, Kondo, J, Yamanaka, D, Kondo, Y, Okamoto, I, Ono, A, Sychrovsky, V, Kojima, C, Tanaka, Y. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of an Ag(I) -Mediated Cytosine-Cytosine Base Pair within DNA Duplex in Solution with (1) H/(15) N/(109) Ag NMR Spectroscopy.
Chemistry, 22, 2016
|
|
5FCL
 
 | | Crystal structure of Cas1 from Pectobacterium atrosepticum | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Wilkinson, M.E, Nakatani, Y, Opel-Reading, H.K, Fineran, P.C, Krause, K.L. | | Deposit date: | 2015-12-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and in vivo activity of Cas1 from the type I-F CRISPR-Cas system.
Biochem.J., 473, 2016
|
|
2ZYZ
 
 | | Pyrobaculum aerophilum splicing endonuclease | | Descriptor: | Putative uncharacterized protein PAE0789, tRNA-splicing endonuclease | | Authors: | Yoshinari, S, Inaoka, D.K, Watanabe, Y, Shiba, T, Kurisu, G, Harada, S. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease
Nucleic Acids Res., 37, 2009
|
|
3A0A
 
 | | Structure of (PPG)4-OPG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Katagiri, A, Tanaka, Y, Nishino, N. | | Deposit date: | 2009-03-13 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of (PPG)4-OPG-(PPG)4
To be Published
|
|
3A92
 
 | | Crystal structure of hen egg white lysozyme soaked with 10mM RhCl3 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
6MWC
 
 | | CryoEM structure of chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-5 antibody | | Descriptor: | E1, E2, EEEV-5 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-29 | | Release date: | 2018-12-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
3VV8
 
 | | Crystal structure of beta secetase in complex with 2-amino-3-methyl-6-((1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl)pyrimidin-4(3H)-one | | Descriptor: | 2-amino-3-methyl-6-[(1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl]pyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
3A95
 
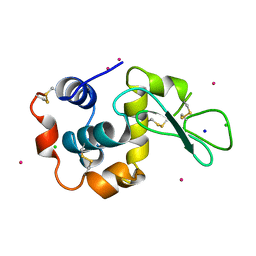 | | Crystal structure of hen egg white lysozyme soaked with 100mM RhCl3 at pH3.8 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
6BQD
 
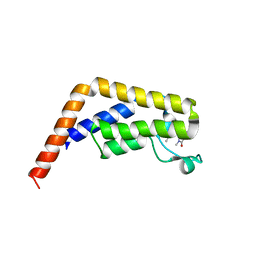 | | TAF1-BD2 bromodomain in complex with (E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
3AF8
 
 | | Crystal Structure of Pd(ally)/apo-C126AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hikage, T, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanism of Accumulation and Incorporation of Organometallic Pd Complexes into the Protein Nanocage of apo-Ferritin.
Inorg.Chem., 49, 2010
|
|
7FGP
 
 | |
5H6O
 
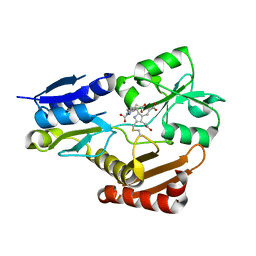 | | Porphobilinogen deaminase from Vibrio Cholerae | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, MAGNESIUM ION, Porphobilinogen deaminase | | Authors: | Funamizu, T, Chen, M, Tanaka, Y, Ishimori, K, Uchida, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Porphobilinogen deaminase from Vibrio Cholerae
To Be Published
|
|
7DO5
 
 | |
3VV7
 
 | | Crystal Structure of beta secetase in complex with 2-amino-6-((1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl)-3-methylpyrimidin-4(3H)-one | | Descriptor: | 2-amino-6-[(1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl]-3-methylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
7DO7
 
 | |
5IJW
 
 | | Glutamate Racemase (MurI) from Mycobacterium smegmatis with bound D-glutamate, 1.8 Angstrom resolution, X-ray diffraction | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase, IODIDE ION | | Authors: | Poen, S, Nakatani, Y, Krause, K. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the structure of glutamate racemase from Mycobacterium tuberculosis as a template for anti-mycobacterial drug discovery.
Biochem. J., 473, 2016
|
|
1MBF
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
2ZUR
 
 | | Crystal Structure of Rh(nbd)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Hirata, K, Ueno, T, Shimizu, N, Yamamoto, M, Takata, M, Watanabe, Y. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polymerization of phenylacetylene by rhodium complexes within a discrete space of apo-ferritin
J.Am.Chem.Soc., 131, 2009
|
|
