5Y4B
 
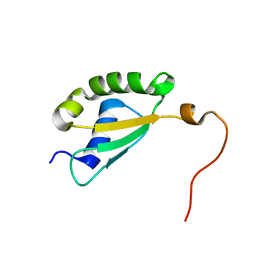 | | Solution structure of yeast Fra2 | | Descriptor: | BolA-like protein 2 | | Authors: | Tang, Y.J, Chi, C.B, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J. Mol. Biol., 430, 2018
|
|
7XXL
 
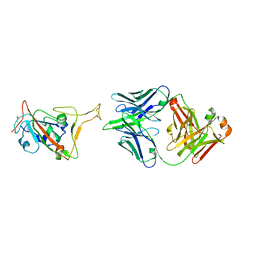 | | RBD in complex with Fab14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | Authors: | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-09-14 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
4A4D
 
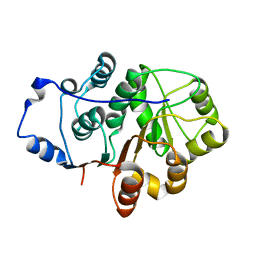 | | Crystal structure of the N-terminal domain of the Human DEAD-BOX RNA helicase DDX5 (P68) | | Descriptor: | PROBABLE ATP-DEPENDENT RNA HELICASE DDX5 | | Authors: | Dutta, S, Choi, Y.W, Kotaka, M, Fielding, B.C, Tan, Y.J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Variable N-Terminal Region of Ddx5 Contains Structural Elements and Auto-Inhibits its Interaction with Ns5B of Hepatitis C Virus.
Biochem.J., 446, 2012
|
|
4AQF
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
4AQG
 
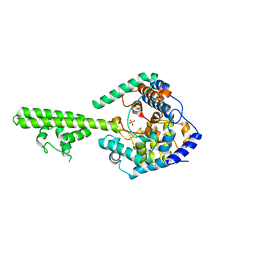 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
2VYI
 
 | | Crystal Structure of the TPR domain of Human SGT | | Descriptor: | SGTA PROTEIN | | Authors: | Dutta, S, Tan, Y.J. | | Deposit date: | 2008-07-24 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Characterization of Human Sgt and its Interaction with Vpu of the Human Immunodeficiency Virus Type 1.
Biochemistry, 47, 2008
|
|
5Y4R
 
 | | Structure of a methyltransferase complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Chemotaxis protein methyltransferase 1, Cyclic diguanosine monophosphate-binding protein PA4608, ... | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
5Y4S
 
 | | Structure of a methyltransferase complex | | Descriptor: | Chemotaxis protein methyltransferase 1 | | Authors: | Yan, X, Xin, L, Tan, Y.J, Jin, S, Liang, Z.X, Gao, Y.G. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Structural analyses unravel the molecular mechanism of cyclic di-GMP regulation of bacterial chemotaxis via a PilZ adaptor protein.
J. Biol. Chem., 293, 2018
|
|
5ZTM
 
 | | Crystal structure of MLE dsRBDs in complex with roX2 (R2H1) | | Descriptor: | Dosage compensation regulator, non-coding mRNA sequence roX2 | | Authors: | Lv, M.Q, Tang, Y.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights reveal the specific recognition of roX RNA by the dsRNA-binding domains of the RNA helicase MLE and its indispensable role in dosage compensation in Drosophila.
Nucleic Acids Res., 47, 2019
|
|
5ZAD
 
 | | Human topoisomerase II beta in complex with DNA | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta | | Authors: | Sun, L.Y, Zhu, L.W, Tang, Y.J. | | Deposit date: | 2018-02-07 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Increasing the distance between two monomers of topoisomerase II beta under the action of antitumor agent 4 beta-sulfur-(benzimidazole) 4'-demethylepipodophyllotoxin.
Sci Rep, 8, 2018
|
|
5Y4U
 
 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | Monothiol glutaredoxin-3 | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y4T
 
 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutaredoxin | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
2FA4
 
 | |
