3AU4
 
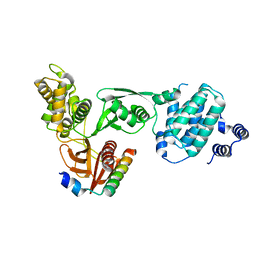 | |
1Z0Q
 
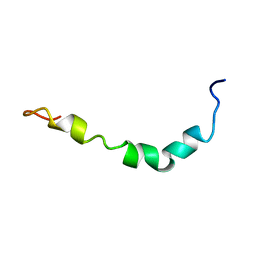 | | Aqueous Solution Structure of the Alzheimer's Disease Abeta Peptide (1-42) | | 分子名称: | Alzheimer's disease amyloid | | 著者 | Tomaselli, S, Esposito, V, Vangone, P, van Nuland, N.A, Bonvin, A.M, Guerrini, R, Tancredi, T, Temussi, P.A, Picone, D. | | 登録日 | 2005-03-02 | | 公開日 | 2006-05-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The alpha-to-beta Conformational Transition of Alzheimer's Abeta-(1-42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of beta Conformation Seeding
Chembiochem, 7, 2006
|
|
1F3Y
 
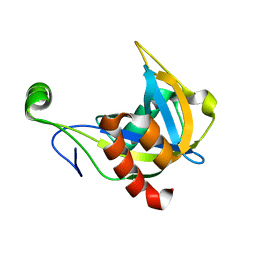 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | 分子名称: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | 著者 | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | 登録日 | 2000-06-06 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
1Z0E
 
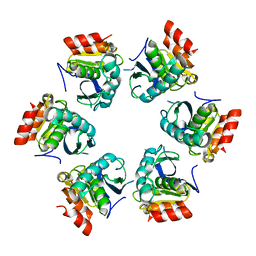 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | 分子名称: | Putative protease La homolog type | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | 登録日 | 2005-03-01 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0C
 
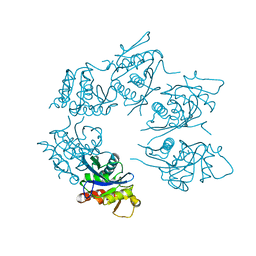 | | Crystal Structure of A. fulgidus Lon proteolytic domain D508A mutant | | 分子名称: | Putative protease La homolog type | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | 登録日 | 2005-03-01 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1Z0G
 
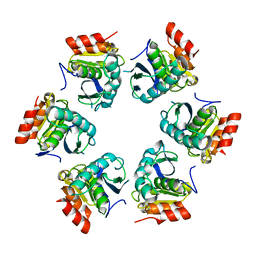 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | 分子名称: | Putative protease La homolog type | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | 登録日 | 2005-03-01 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
2ANE
 
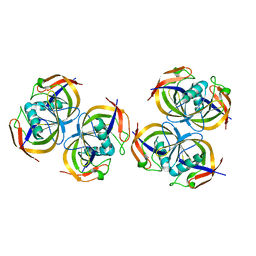 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | 分子名称: | ATP-dependent protease La | | 著者 | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | 登録日 | 2005-08-11 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
2QUL
 
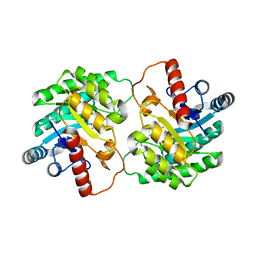 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii at 1.79 A resolution | | 分子名称: | D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUM
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii with D-tagatose | | 分子名称: | D-tagatose, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUN
 
 | | Crystal Structure of D-tagatose 3-epimerase from Pseudomonas cichorii in Complex with D-fructose | | 分子名称: | D-fructose, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | 登録日 | 2007-08-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | 分子名称: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | 著者 | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | 登録日 | 2015-07-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | 分子名称: | ATP-dependent protease La, SULFATE ION | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | 登録日 | 2003-12-08 | | 公開日 | 2003-12-23 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
2CYM
 
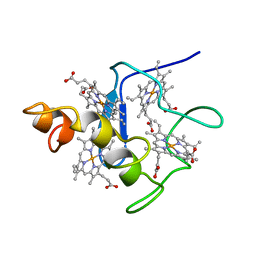 | | EFFECTS OF AMINO ACID SUBSTITUTION ON THREE-DIMENSIONAL STRUCTURE: AN X-RAY ANALYSIS OF CYTOCHROME C3 FROM DESULFOVIBRIO VULGARIS HILDENBOROUGH AT 2 ANGSTROMS RESOLUTION | | 分子名称: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Morimoto, Y, Tani, T, Okumura, H, Higuchi, Y, Yasuoka, N. | | 登録日 | 1993-09-29 | | 公開日 | 1994-04-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of amino acid substitution on three-dimensional structure: an X-ray analysis of cytochrome c3 from Desulfovibrio vulgaris Hildenborough at 2 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1QZM
 
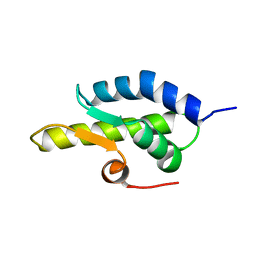 | | alpha-domain of ATPase | | 分子名称: | ATP-dependent protease La | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Khalatova, A.G, Rasulova, F.S, Tropea, J.E, Maurizi, M.R, Rotanova, T.V, Gustchina, A, Wlodawer, A. | | 登録日 | 2003-09-17 | | 公開日 | 2004-05-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the AAA+ alpha domain of E. coli Lon protease at 1.9A resolution.
J.Struct.Biol., 146
|
|
6IBI
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | 著者 | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | 登録日 | 2018-11-30 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
5YAE
 
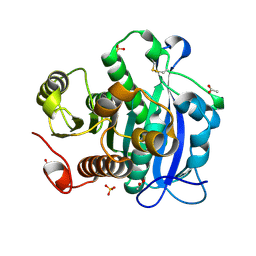 | | Ferulic acid esterase from Streptomyces cinnamoneus at 2.4 A resolution | | 分子名称: | ACETATE ION, Esterase, SULFATE ION | | 著者 | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | 登録日 | 2017-08-31 | | 公開日 | 2017-12-06 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
1RRE
 
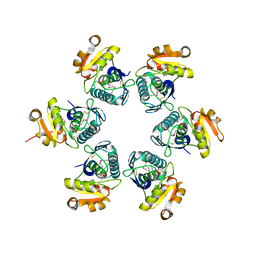 | | Crystal structure of E.coli Lon proteolytic domain | | 分子名称: | ATP-dependent protease La, SULFATE ION | | 著者 | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | 登録日 | 2003-12-08 | | 公開日 | 2004-02-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | 分子名称: | ACETATE ION, Esterase, GLYCEROL, ... | | 著者 | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | 登録日 | 2017-09-01 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
1KZT
 
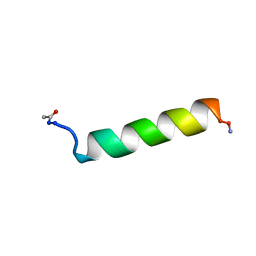 | |
1KZS
 
 | |
1KZV
 
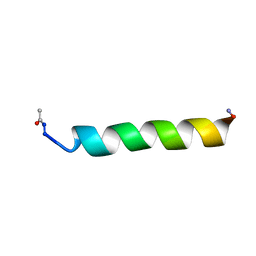 | |
1M9G
 
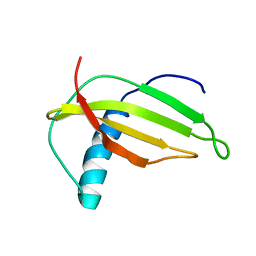 | | Solution structure of G16A-MNEI, a structural mutant of single chain monellin MNEI | | 分子名称: | Monellin chain B and Monellin chain A | | 著者 | Spadaccini, R, Trabucco, F, Saviano, G, Picone, D, Crescenzi, O, Tancredi, T, Temussi, P.A. | | 登録日 | 2002-07-29 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Mechanism of Interaction of Sweet Proteins with the T1R2-T1R3 Receptor: Evidence from the Solution Structure of G16A-MNEI
J.MOL.BIOL., 328, 2003
|
|
7DAJ
 
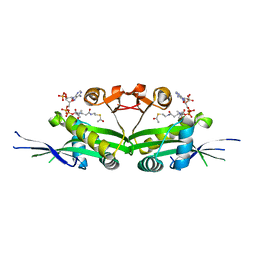 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | 分子名称: | Serotonin N-acetyltransferase 1, chloroplastic | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAL
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | 分子名称: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | 著者 | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | 登録日 | 2020-10-16 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
