5FD9
 
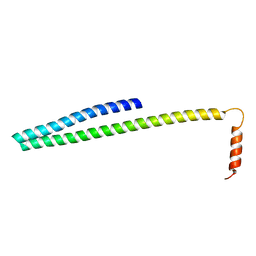 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation B) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
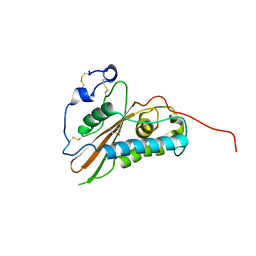
2VZN
 
 | |
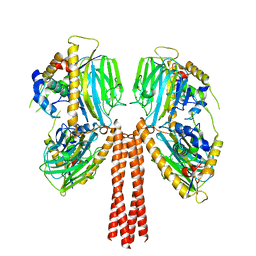
8JP6
 
 | |
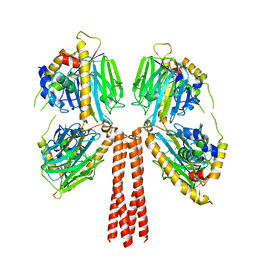
8JPG
 
 | | Cryo-EM structure of full-length ERGIC-53 with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53, ... | | Authors: | Watanabe, S, Inaba, K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | Structure of full-length ERGIC-53 in complex with MCFD2 for cargo transport.
Nat Commun, 15, 2024
|
|
8JP7
 
 | |
8JP8
 
 | |
8JP9
 
 | |
8JP5
 
 | |
8JP4
 
 | |
2W27
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | Authors: | Padavattan, S, AndERSON, W.F, Schirmer, T. | | Deposit date: | 2008-10-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
5FD7
 
 | | X-ray Crystal Structure of ESCRT-III Snf7 core domain (conformation A) | | Descriptor: | Vacuolar-sorting protein SNF7 | | Authors: | Tang, S, Henne, W.M, Borbat, P.P, Buchkovich, N.J, Freed, J.H, Mao, Y, Fromme, J.C, Emr, S.D. | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments.
Elife, 4, 2015
|
|
1U7Z
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, 4'-phosphopantothenoyl-CMP complex | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7U
 
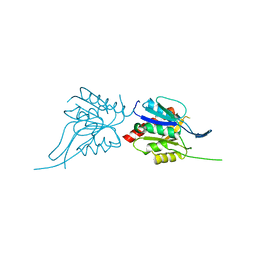 | | Phosphopantothenoylcysteine synthetase from E. coli | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7W
 
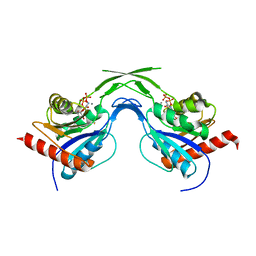 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
2QW7
 
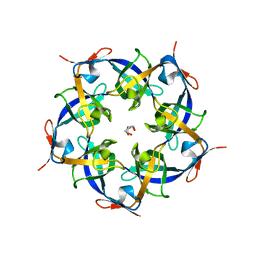 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
5AYK
 
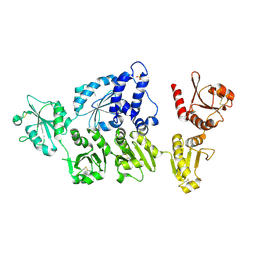 | | Crystal structure of ERdj5 form I | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
5AYL
 
 | | Crystal structure of ERdj5 form II | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
6CYJ
 
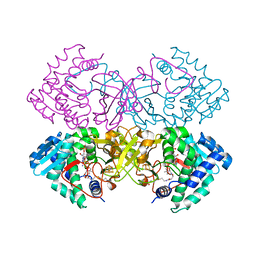 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER, SUCCINYL-COENZYME A | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
6CZ6
 
 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | COENZYME A, HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
6D2S
 
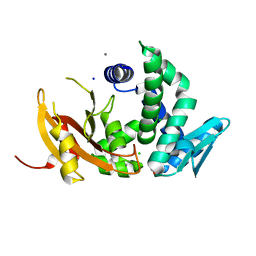 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-13 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
7JST
 
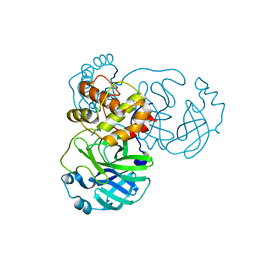 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT0
 
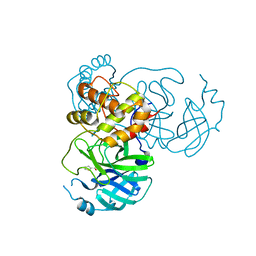 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
6CYY
 
 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | COENZYME A, HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
7JW8
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 in space group P1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
