2Z44
 
 | | Crystal Structure of Selenomethionine-labeled ORF134 | | Descriptor: | ORF134 | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
3AFG
 
 | | Crystal structure of ProN-Tk-SP from Thermococcus kodakaraensis | | Descriptor: | CALCIUM ION, Subtilisin-like serine protease | | Authors: | Foophow, T, Tanaka, S, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin homologue, Tk-SP, from Thermococcus kodakaraensis: requirement of a C-terminal beta-jelly roll domain for hyperstability.
J.Mol.Biol., 400, 2010
|
|
2EYX
 
 | |
2EYV
 
 | | SH2 domain of CT10-Regulated Kinase | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2EYW
 
 | |
2EYY
 
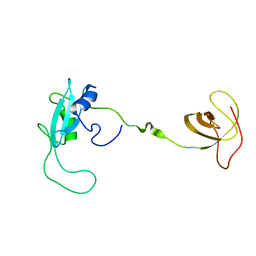 | | CT10-Regulated Kinase isoform I | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1U9I
 
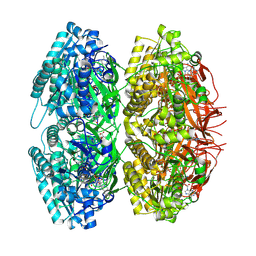 | | Crystal Structure of Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC, MAGNESIUM ION | | Authors: | Xu, Y, Mori, T, Pattanayek, R, Pattanayek, S, Egli, M, Johnson, C.H. | | Deposit date: | 2004-08-09 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
7DVE
 
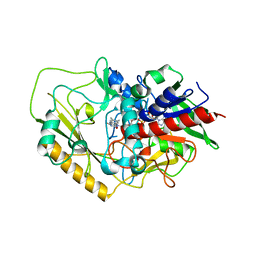 | | Crystal structure of FAD-dependent C-glycoside oxidase | | Descriptor: | 6'''-hydroxyparomomycin C oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Senda, M, Watanabe, S, Kumano, T, Kobayashi, M, Senda, T. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FAD-dependent C -glycoside-metabolizing enzymes in microorganisms: Screening, characterization, and crystal structure analysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BUX
 
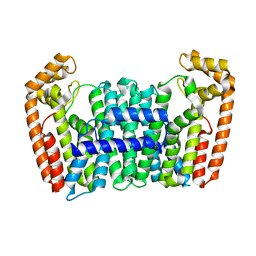 | | Eucommia ulmoides FPS1 | | Descriptor: | FPS2 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUW
 
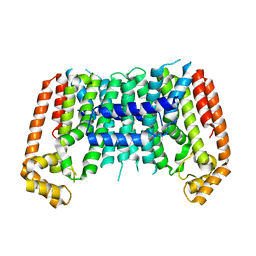 | | Eucommia ulmoides TPT3 mutant -C94Y/A95F | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUV
 
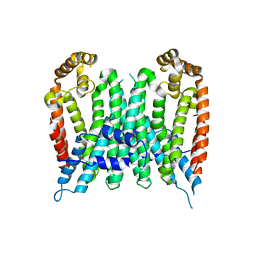 | | Eucommia ulmoides TPT3, crystal form 2 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
7BUU
 
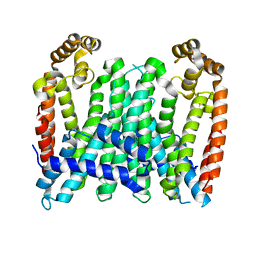 | | Eucommia ulmoides TPT3, crystal form 1 | | Descriptor: | FPS3 | | Authors: | Kajiura, H, Yoshizawa, T, Tokumoto, Y, Suzuki, N, Takeno, S, Takeno, K.J, Yamashita, T, Tanaka, S, Kaneko, Y, Fujiyama, K, Matsumura, H, Nakazawa, Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of ultrahigh molecular weight isoprenes provide key insights into their biosynthesis.
Commun Biol, 4, 2021
|
|
6JJU
 
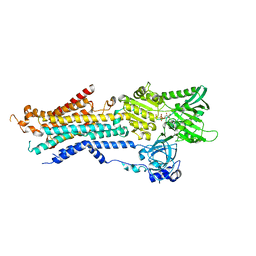 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
1XMF
 
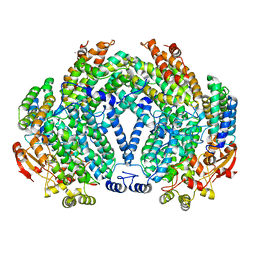 | | Structure of Mn(II)-Soaked Apo Methane Monooxygenase Hydroxylase Crystals from M. capsulatus (Bath) | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Methane monooxygenase component A alpha chain, ... | | Authors: | Sazinsky, M.H, Merkx, M, Cadieux, E, Tang, S, Lippard, S.J. | | Deposit date: | 2004-10-02 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Preparation and X-ray Structures of Metal-Free, Dicobalt and Dimanganese Forms of Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
1D0E
 
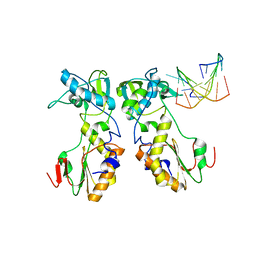 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
2Z6V
 
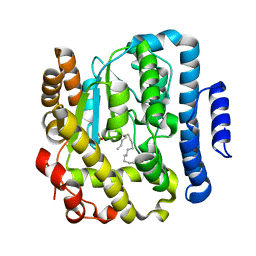 | | Crystal structure of sulfotransferase STF9 from Mycobacterium avium | | Descriptor: | PALMITIC ACID, Putative uncharacterized protein, SULFATE ION | | Authors: | Hassain, M.M, Moriizumi, Y, Tanaka, S, Kimura, M, Kakuta, Y. | | Deposit date: | 2007-08-09 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of sulfotransferase STF9 from Mycobacterium avium
To be Published
|
|
2KUQ
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
1YTV
 
 | | Maltose-binding protein fusion to a C-terminal fragment of the V1a vasopressin receptor | | Descriptor: | Maltose-binding periplasmic protein, Vasopressin V1a receptor, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Adikesavan, N.V, Mahmood, S.S, Stanley, S, Xu, Z, Wu, N, Thibonnier, M, Shoham, M. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-terminal segment of the V1R vasopressin receptor is unstructured in the crystal structure of its chimera with the maltose-binding protein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
2B5S
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2ALG
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | Descriptor: | AVR3a4 | | Authors: | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | Deposit date: | 2011-04-12 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
2QKE
 
 | |
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
