6X6A
 
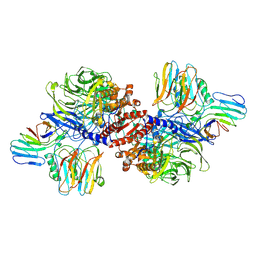 | | Cryo-EM structure of NLRP1-DPP9 complex | | 分子名称: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1 | | 著者 | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | 登録日 | 2020-05-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
8F4V
 
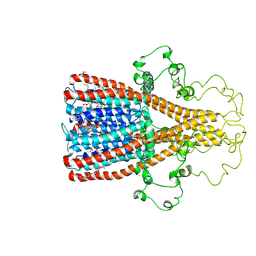 | | Alpha7 nicotinic acetylcholine receptor intracellular and transmembrane domains bound to ivermectin in a desensitized state | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Neuronal acetylcholine receptor subunit alpha-7 | | 著者 | Bondarenko, V, Chen, Q, Tang, P. | | 登録日 | 2022-11-11 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Elucidation of Ivermectin Binding to alpha 7nAChR and the Induced Channel Desensitization.
Acs Chem Neurosci, 14, 2023
|
|
6XKJ
 
 | | Cryo-EM structure of CARD8-CARD filament | | 分子名称: | Caspase recruitment domain-containing protein 8 | | 著者 | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | 登録日 | 2020-06-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
6GF3
 
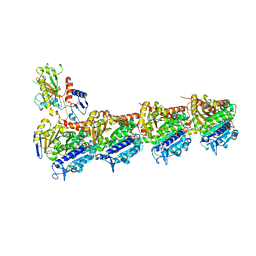 | | Tubulin-Jerantinine B acetate complex | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Smedley, C.J, Stanley, P.A, Qazzaz, M.E, Prota, A.E, Olieric, N, Collins, H, Eastman, H, Barrow, A.S, Lim, K.-H, Kam, T.-S, Smith, B.J, Duivenvoorden, H.M, Parker, B.S, Bradshaw, T.D, Steinmetz, M.O, Moses, J.E. | | 登録日 | 2018-04-29 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Sustainable Syntheses of (-)-Jerantinines A & E and Structural Characterisation of the Jerantinine-Tubulin Complex at the Colchicine Binding Site.
Sci Rep, 8, 2018
|
|
7ZRM
 
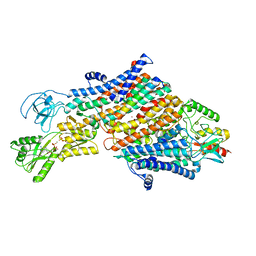 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRD
 
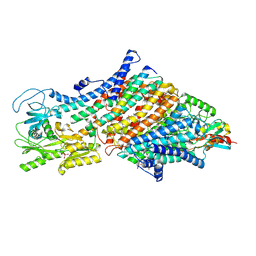 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
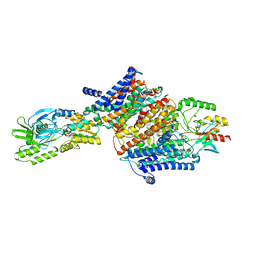 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | 分子名称: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRE
 
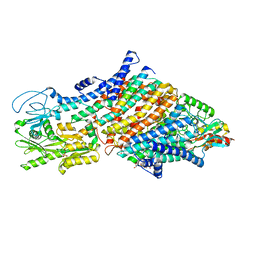 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, under turnover conditions | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
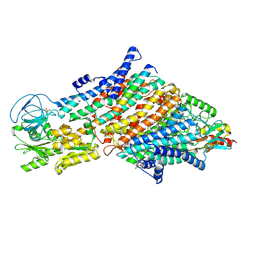 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRJ
 
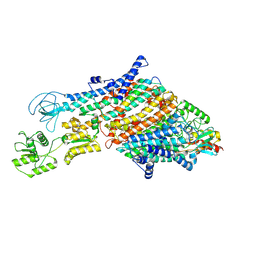 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRG
 
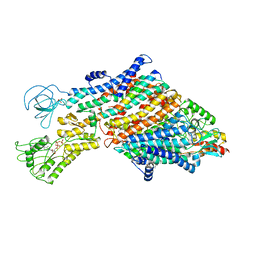 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRI
 
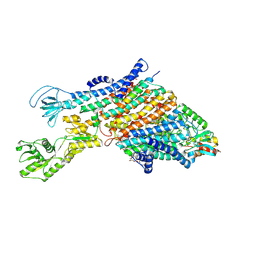 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | 分子名称: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
2M6I
 
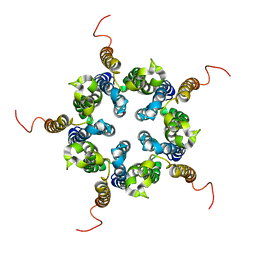 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-29 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
2M6B
 
 | | Structure of full-length transmembrane domains of human glycine receptor alpha1 monomer subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-28 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
5OON
 
 | | Structure of Undecaprenyl-Pyrophosphate Phosphatase, BacA | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | 著者 | Huang, C.-Y, Olieric, V, Warshamanage, R, Wang, M, Howe, N, Ghachi, M.E.I, Weichert, D, Kerff, F, Stansfeld, P, Touze, T, Caffrey, M. | | 登録日 | 2017-08-08 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.
Nat Commun, 9, 2018
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | 分子名称: | COENZYME F420, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | 分子名称: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Ariza, A, Fontana, P. | | 登録日 | 2022-07-08 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.508 Å) | | 主引用文献 | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
1MOT
 
 | | NMR Structure Of Extended Second Transmembrane Domain Of Glycine Receptor alpha1 Subunit in SDS Micelles | | 分子名称: | Glycine Receptor alpha-1 CHAIN | | 著者 | Yushmanov, V.E, Mandal, P.K, Liu, Z, Tang, P, Xu, Y. | | 登録日 | 2002-09-09 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure and Backbone Dynamics of the Extended Second Transmembrane Domain of the Human Neuronal Glycine Receptor Alpha1 Subunit
Biochemistry, 42, 2003
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | 分子名称: | COENZYME F420, Putative uncharacterized protein | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Z
 
 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | 分子名称: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | 分子名称: | COENZYME F420, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | 分子名称: | Deazaflavin-dependent nitroreductase, SULFATE ION | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3PJR
 
 | | HELICASE SUBSTRATE COMPLEX | | 分子名称: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | 登録日 | 1999-03-12 | | 公開日 | 1999-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism
Cell(Cambridge,Mass.), 97, 1999
|
|
4I5M
 
 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | 分子名称: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | 著者 | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | 登録日 | 2012-11-28 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
