5IAU
 
 | |
5INX
 
 | |
6LL8
 
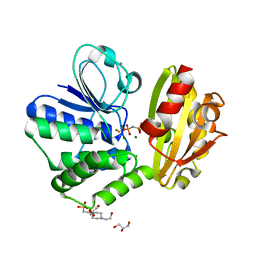 | | Type II inorganic pyrophosphatase (PPase) from the psychrophilic bacterium Shewanella sp. AS-11, Mg-PNP form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, FLUORIDE ION, ... | | Authors: | Horitani, M, Kusubayashi, K, Oshima, K, Yato, A, Sugimoto, H, Watanabe, K. | | Deposit date: | 2019-12-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystallography and Electron Paramagnetic Resonance Spectroscopy Reveal Active Site Rearrangement of Cold-Adapted Inorganic Pyrophosphatase.
Sci Rep, 10, 2020
|
|
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4UX1
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
7YRK
 
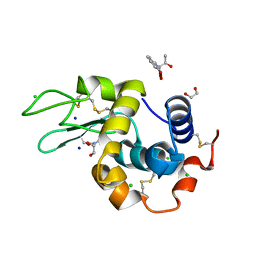 | | Crystal structure of the hen egg lysozyme-2-oxidobenzylidene-threoninato-copper (II) complex | | Descriptor: | (6~{S})-6-[(1~{R})-1-oxidanylethyl]-2,4-dioxa-7$l^{4}-aza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(13),7,9,11-tetraen-5-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Unno, M, Furuya, T, Kitanishi, K, Akitsu, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Sci Rep, 13, 2023
|
|
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
7DQE
 
 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7DQC
 
 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
1QA9
 
 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | Descriptor: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | Authors: | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | Deposit date: | 1999-04-13 | | Release date: | 1999-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
3VLE
 
 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLD
 
 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLF
 
 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VL1
 
 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4YE5
 
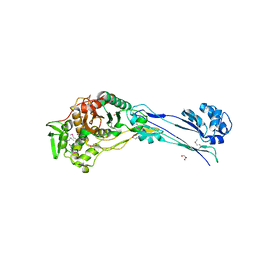 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
3KDQ
 
 | | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae. | | Descriptor: | uncharacterized conserved protein | | Authors: | Zhang, R, Wu, R, Tan, K, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a functionally unknown conserved protein from Corynebacterium diphtheriae.
To be Published
|
|
4XQ2
 
 | | Ensemble refinement of cystathione gamma lyase (CalE6) D7G from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, CalE6, ... | | Authors: | Wang, F, Yennamalli, R.M, Singh, S, Tan, K, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora
To Be Published
|
|
3VR2
 
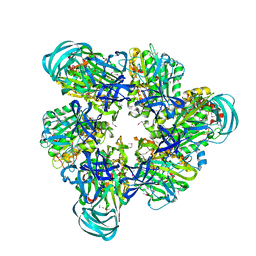 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR4
 
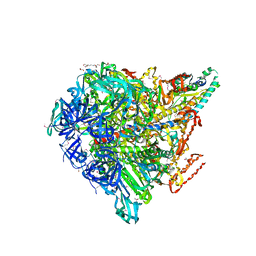 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
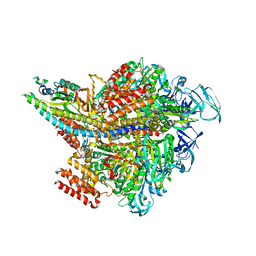 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR5
 
 | | Crystal structure of nucleotide-free Enterococcus hirae V1-ATPase [eV1(L)] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B, V-type sodium ATPase subunit D, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
2RRD
 
 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
3OOS
 
 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
