4NPB
 
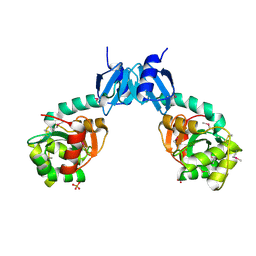 | | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92 | | Descriptor: | PHOSPHATE ION, Protein disulfide isomerase II, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92
To be Published
|
|
8ZRT
 
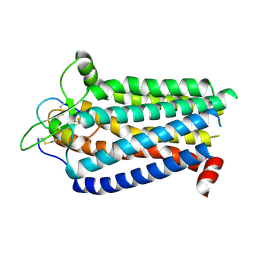 | | Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1 | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
8XWP
 
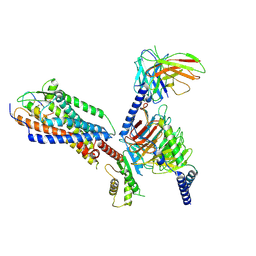 | | Cryo-EM structure of ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
8XWQ
 
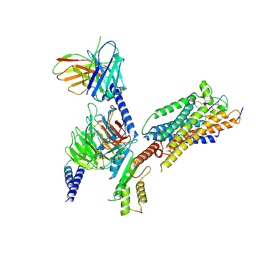 | | Cryo-EM structure of ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
5CD2
 
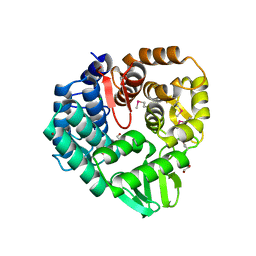 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | Descriptor: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-02 | | Release date: | 2015-07-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
7KYU
 
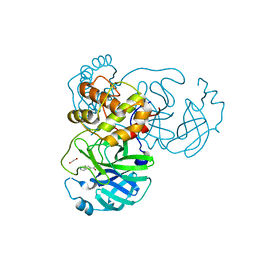 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
4NOC
 
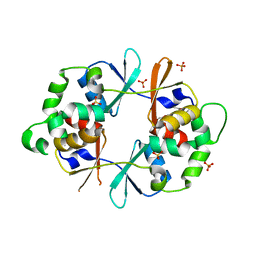 | |
4NEG
 
 | | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | FORMIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4Q6T
 
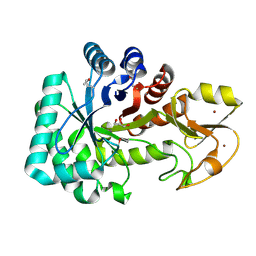 | | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5 | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structure of a class V chitininase from Pseudomonas fluorescens Pf-5
To be Published
|
|
4PYS
 
 | | The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343 | | Descriptor: | FORMIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | The crystal structure of beta-N-acetylhexosaminidase from Bacteroides fragilis NCTC 9343
To be Published
|
|
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
4Q7Q
 
 | | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Lipolytic protein G-D-S-L family, ... | | Authors: | Tan, K, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588
To be Published
|
|
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
5D5H
 
 | | Crystal structure of Mycobacterium tuberculosis Topoisomerase I | | Descriptor: | ACETATE ION, DNA topoisomerase 1, GLYCEROL, ... | | Authors: | Tan, K, Cheng, B, Tse-Dinh, Y.C. | | Deposit date: | 2015-08-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold.
J.Mol.Biol., 428, 2016
|
|
4Q7O
 
 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | Descriptor: | BROMIDE ION, FORMIC ACID, Immunity protein | | Authors: | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4Q2B
 
 | | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-D-glucanase, FORMIC ACID, ... | | Authors: | Tan, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440
To be Published
|
|
5EV7
 
 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
4NQR
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with alanine | | Descriptor: | ALANINE, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with alanine.
To be Published
|
|
4DQD
 
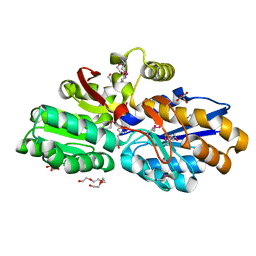 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4O5A
 
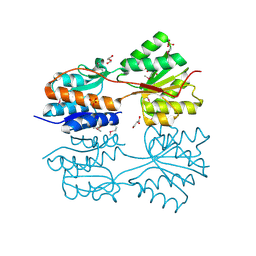 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4NZP
 
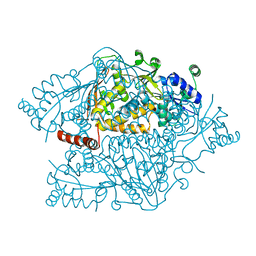 | | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Argininosuccinate synthase | | Authors: | Tan, K, Gu, M, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | The crystal structure of argininosuccinate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4OAT
 
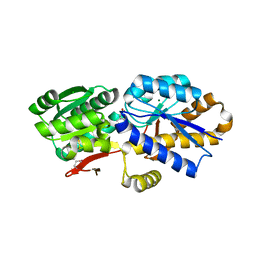 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with isoleucine. | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-06 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with isoleucine.
To be Published
|
|
4OBB
 
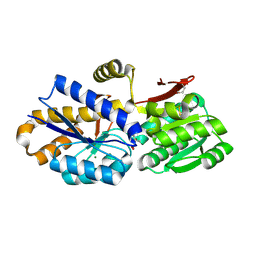 | | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid. | | Descriptor: | (3S)-3-methyl-2-oxopentanoic acid, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | The crystal structure of a solute-binding protein from Anabaena variabilis ATCC 29413 in complex with (3S)-3-methyl-2-oxopentanoic acid.
To be Published
|
|
4OTZ
 
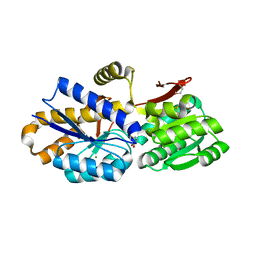 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CYSTEINE, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with cystein
To be Published
|
|
