8WJ2
 
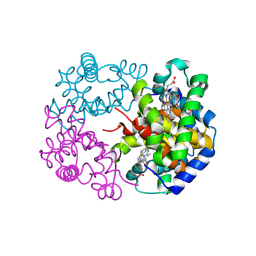 | | cryo-EM structure of human haemoglobin in deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8WJ5
 
 | | Double-wing domain of E. coli YdcD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Uncharacterized protein YdcD | | Authors: | Kamata, K, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making the unphasable feasible
To Be Published
|
|
5F53
 
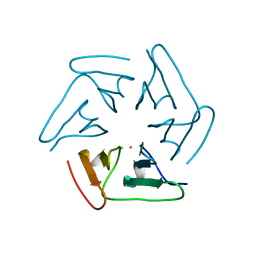 | |
1RKM
 
 | | STRUCTURE OF OPPA | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
8Y5V
 
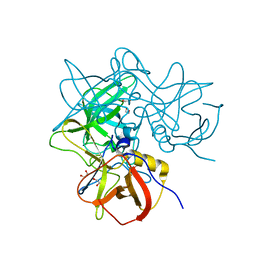 | | GII.4 Sydney PD and 2'-FL | | Descriptor: | 1,2-ETHANEDIOL, GII.4 Sydney PD in complex with 2'-FL (powder), alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | GII.4 Sydney PD and 2'-FL
To Be Published
|
|
8Y6C
 
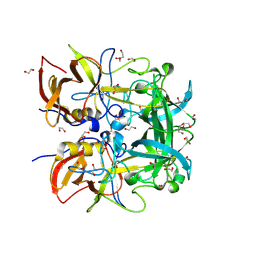 | | Norovirus GII.10 P domain and 2'-FL (powder) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'FL (powder), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Norovirus GII.10 P domain and 2'-FL (powder)
To Be Published
|
|
8Y6D
 
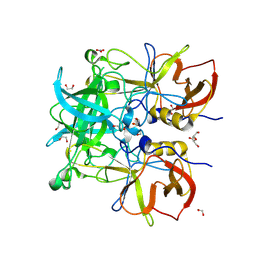 | | Norovirus GII.10 P domain and 2'-FL (tablet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'-FL (tablet), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Norovirus GII.10 P domain and 2'-FL (tablet)
To Be Published
|
|
5CHB
 
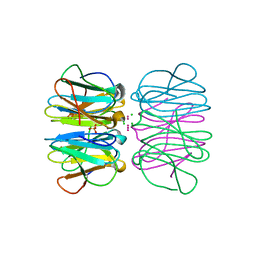 | | Crystal structure of nvPizza2-S16H58 coordinating a CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ZCN
 
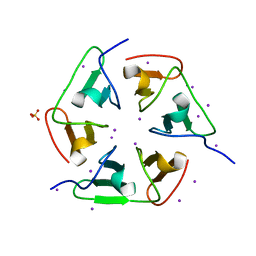 | | Crystal structure of nvPizza2-S16S58 | | Descriptor: | IODIDE ION, SULFATE ION, nvPizza2-S16S58 | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of nvPizza2-S16S58
To Be Published
|
|
1WUB
 
 | | Crystal structure of the polyisoprenoid-binding protein, TT1927b, from Thermus thermophilus HB8 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, conserved hypothetical protein TT1927b | | Authors: | Handa, N, Idaka, M, Terada, T, Hamana, H, Ishizuka, Y, Park, S.-Y, Tame, J.R.H, Doi-Katayama, Y, Hirota, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8
Protein Sci., 14, 2005
|
|
3X1Y
 
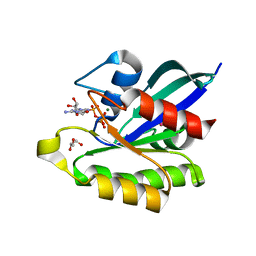 | | Ras-related protein Rap1B(L9V) with GppNHp | | Descriptor: | CADMIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Noguchi, H, Ikegami, T, Park, S.Y, Tame, J.R.H, Unzai, S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | The structure and conformational switching of Rap1B
Biochem.Biophys.Res.Commun., 462, 2015
|
|
3X1W
 
 | | Ras-related protein Rap1B with GDP | | Descriptor: | CADMIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Noguchi, H, Ikegami, T, Akashi, S, Park, S.Y, Tame, J.R.H, Unzai, S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure and conformational switching of Rap1B
Biochem.Biophys.Res.Commun., 462, 2015
|
|
3X1X
 
 | | Ras-related protein Rap1B with GppNHp | | Descriptor: | CADMIUM ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Noguchi, H, Ikegami, T, Park, S.Y, Tame, J.R.H, Unzai, S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The structure and conformational switching of Rap1B
Biochem.Biophys.Res.Commun., 462, 2015
|
|
3X1Z
 
 | | Ras-related protein Rap1B(T65A) with GppNHp | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Noguchi, H, Ikegami, T, Park, S.Y, Tame, J.R.H, Unzai, S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-06-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The structure and conformational switching of Rap1B
Biochem.Biophys.Res.Commun., 462, 2015
|
|
1UIU
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIV
 
 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIW
 
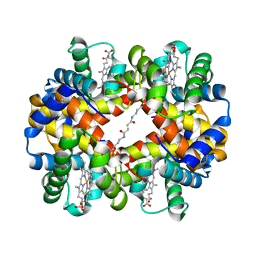 | | Crystal Structures of Unliganded and Half-Liganded Human Hemoglobin Derivatives Cross-Linked between Lys 82beta1 and Lys 82beta2 | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Park, S.-Y, Shibayama, N, Tame, J.R.H. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of unliganded and half-liganded human hemoglobin derivatives cross-linked between Lys 82beta1 and Lys 82beta2
Biochemistry, 43, 2004
|
|
1WXR
 
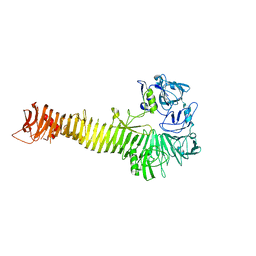 | | Crystal structure of Heme Binding protein, an autotransporter hemoglobine protease from pathogenic Escherichia coli | | Descriptor: | haemoglobin protease | | Authors: | Otto, B.R, Sijbrandi, R, Luirink, J, Oudega, B, Heddle, J.G, Mizutani, K, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic escherichia coli
J.Biol.Chem., 280, 2005
|
|
3VUS
 
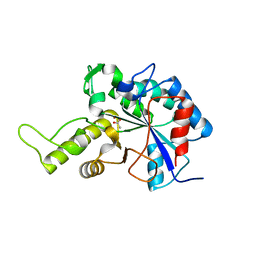 | | Escherichia coli PgaB N-terminal domain | | Descriptor: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-07-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1V9F
 
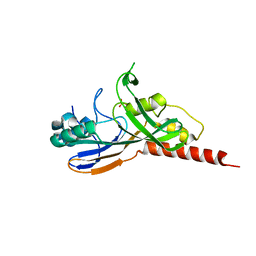 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1V9K
 
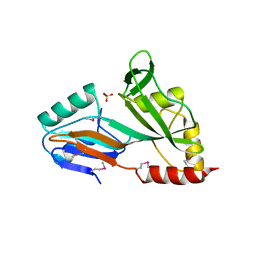 | | The crystal structure of the catalytic domain of pseudouridine synthase RluC from Escherichia coli | | Descriptor: | Ribosomal large subunit pseudouridine synthase C, SULFATE ION | | Authors: | Machida, Y, Mizutani, K, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
3WW7
 
 | | Crystal structure of the computationally designed Pizza2 protein | | Descriptor: | GLYCEROL, Pizza2 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMU
 
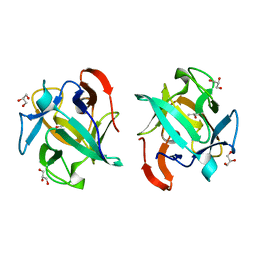 | | The structure of an anti-cancer lectin mytilec apo-form from the mussel Mytilus galloprovincialis | | Descriptor: | GLYCEROL, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
3WW8
 
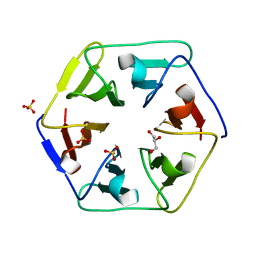 | | Crystal structure of the computationally designed Pizza3 protein | | Descriptor: | GLYCEROL, Pizza3 protein, SULFATE ION | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
