8Q5I
 
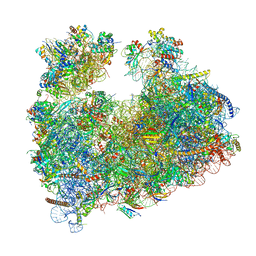 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | 分子名称: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
6MJU
 
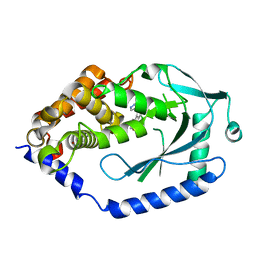 | | human cGAS catalytic domain bound with the inhibitor G108 | | 分子名称: | 1-[6,7-dichloro-9-(1H-pyrazol-4-yl)-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | 登録日 | 2018-09-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
4EFM
 
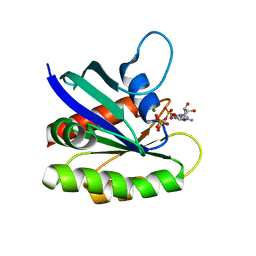 | | Crystal structure of H-Ras G12V in complex with GppNHp (state 1) | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | 登録日 | 2012-03-30 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | 著者 | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | 登録日 | 2014-03-26 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
3VSR
 
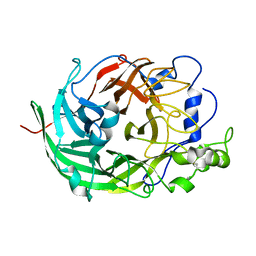 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | 分子名称: | Beta-fructofuranosidase | | 著者 | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | 登録日 | 2012-05-08 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSS
 
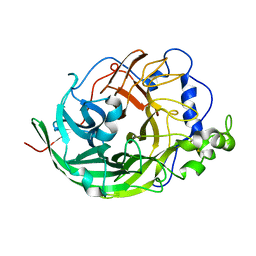 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | 分子名称: | Beta-fructofuranosidase, beta-D-fructofuranose | | 著者 | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | 登録日 | 2012-05-08 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3ZLF
 
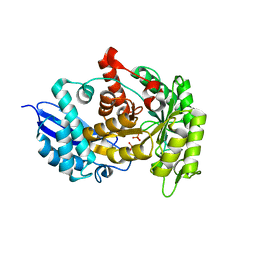 | | Structure of group A Streptococcal enolase K312A mutant | | 分子名称: | ENOLASE, PHOSPHATE ION | | 著者 | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | 登録日 | 2013-01-31 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
1ITP
 
 | | Solution Structure of POIA1 | | 分子名称: | proteinase A inhibitor 1 | | 著者 | Sasakawa, H, Yoshinaga, S, Kojima, S, Tamura, A. | | 登録日 | 2002-01-23 | | 公開日 | 2002-02-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of POIA1, a homologous protein to the propeptide of subtilisin: implication for protein foldability and the function as an intramolecular chaperone.
J.Mol.Biol., 317, 2002
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | 分子名称: | Claudin-19, Heat-labile enterotoxin B chain | | 著者 | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | 登録日 | 2014-12-13 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
