3WOA
 
 | |
6JGJ
 
 | | Crystal structure of the F99S/M153T/V163A/E222Q variant of GFP at 0.78 A | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Takaba, K, Tai, Y, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
8WU5
 
 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
5D8V
 
 | | Ultra-high resolution structure of high-potential iron-sulfur protein | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Hirano, Y, Takeda, K, Miki, K. | | Deposit date: | 2015-08-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.48 Å) | | Cite: | Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48 angstrom
Nature, 534, 2016
|
|
5DHE
 
 | | Crystal structure of ChBD3 from Thermococcus kodakarensis KOD1 | | Descriptor: | Chitinase, GLYCEROL | | Authors: | Niwa, S, Hibi, M, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
6IED
 
 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CXM
 
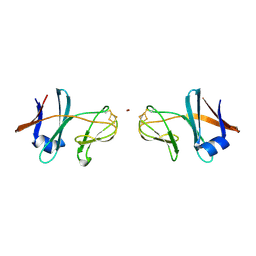 | | Crystal structure of the cyanobacterial plasma membrane Rieske protein PetC3 from Synechocystis PCC 6803 | | Descriptor: | Cytochrome b6/f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER, NICKEL (II) ION, ... | | Authors: | Veit, S, Takeda, K, Miki, K, Roegner, M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterisation of the cyanobacterial PetC3 Rieske protein family.
Biochim. Biophys. Acta, 1857, 2016
|
|
4V8K
 
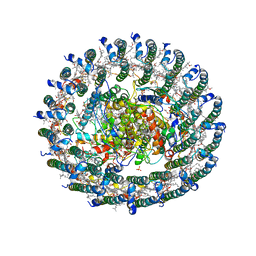 | | Crystal structure of the LH1-RC complex from Thermochromatium tepidum in P21 form | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CALCIUM ION, ... | | Authors: | Niwa, S, Takeda, K, Wang-Otomo, Z.-Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 angstrom
Nature, 508, 2014
|
|
3FF5
 
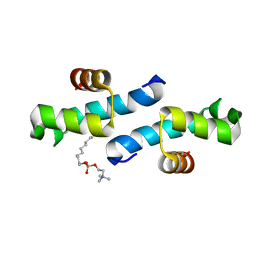 | | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix-protein-import receptor, Pex14p | | Descriptor: | Peroxisomal biogenesis factor 14, decyl 2-trimethylazaniumylethyl phosphate | | Authors: | Su, J.-R, Takeda, K, Tamura, S, Fujiki, Y, Miki, K. | | Deposit date: | 2008-12-01 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HRX
 
 | |
3M0V
 
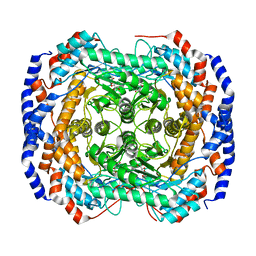 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
7VOS
 
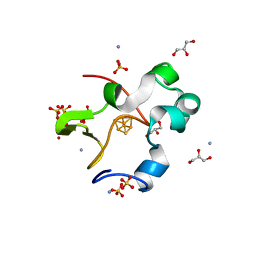 | | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | AMMONIUM ION, GLYCEROL, High-potential iron-sulfur protein, ... | | Authors: | Hanazono, Y, Hirano, Y, Takeda, K, Kusaka, K, Tamada, T, Miki, K. | | Deposit date: | 2021-10-14 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (0.66 Å), X-RAY DIFFRACTION | | Cite: | Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis.
Sci Adv, 8, 2022
|
|
3M0X
 
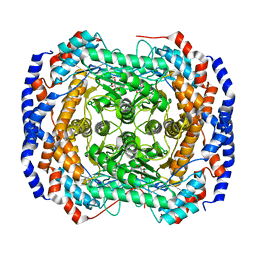 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0M
 
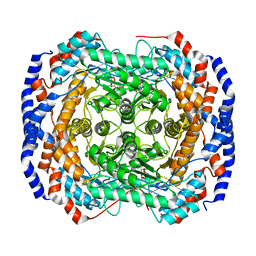 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0H
 
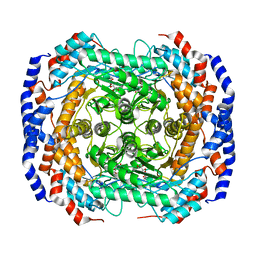 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0Y
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329A in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0L
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7XJC
 
 | | Crystal structure of bacteriorhodopsin in the ground and K states after green laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJE
 
 | | Crystal structure of bacteriorhodopsin in the K state refined against the extrapolated dataset | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin
Commun Biol, 6, 2023
|
|
7XJD
 
 | | Crystal structure of bacteriorhodopsin in the ground state by red laser irradiation | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Taguchi, S, Niwa, S, Takeda, K. | | Deposit date: | 2022-04-16 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Detailed analysis of distorted retinal and its interaction with surrounding residues in the K intermediate of bacteriorhodopsin.
Commun Biol, 6, 2023
|
|
7YRA
 
 | | Crystal structure of [2Fe-2S]-TtPetA | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Tsutsumi, E, Niwa, S, Takeda, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.
Commun Chem, 6, 2023
|
|
7YR9
 
 | | Crystal structure of the immature form of TtPetA | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tsutsumi, E, Niwa, S, Takeda, K. | | Deposit date: | 2022-08-09 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative immature form of a Rieske-type iron-sulfur protein in complex with zinc chloride.
Commun Chem, 6, 2023
|
|
5GV8
 
 | |
5GV7
 
 | |
