9ATI
 
 | | Crystal structure of MERS 3CL protease in complex with a racemic bicyclo[2.2.1]heptenyl-methyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ASW
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATH
 
 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9BK2
 
 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BK3
 
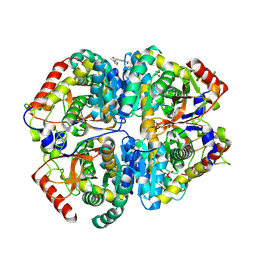 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BTS
 
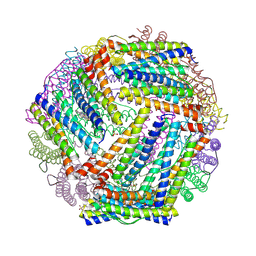 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
6NLN
 
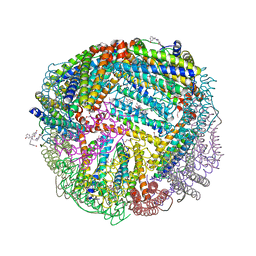 | | 1.60 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 16) | | Descriptor: | 4-{[3-(3-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLJ
 
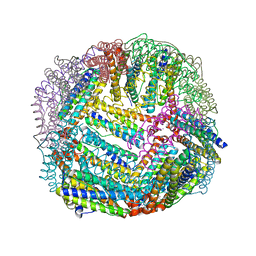 | | 1.65 A resolution structure of Apo BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 12) | | Descriptor: | 4-{[(3-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLF
 
 | | 1.45 A resolution structure of apo BfrB from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLL
 
 | | 1.80 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 14) | | Descriptor: | 4-{[3-(2,4-dihydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
8W1F
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer, Mg bound) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
4XBD
 
 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
4XBC
 
 | | 1.60 A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Hexagonal Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
4XBB
 
 | | 1.85A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Descriptor: | 3C-LIKE PROTEASE, SULFATE ION, diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
6NLI
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 11) | | Descriptor: | 4-{[(2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLM
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 15) | | Descriptor: | 4-{[3-(4-hydroxy-2-methoxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLG
 
 | | 1.50 A resolution structure of BfrB (C89S/K96C) from Pseudomonas aeruginosa in complex with a small molecule fragment (analog 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-hydroxy-1H-isoindole-1,3(2H)-dione, Bacterioferritin, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLK
 
 | | 1.85 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 13) | | Descriptor: | 4-{[3-(4-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6XMK
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6MAB
 
 | | 1.7A resolution structure of RsbU from Chlamydia trachomatis (periplasmic domain) | | Descriptor: | ISOPROPYL ALCOHOL, Sigma regulatory family protein-PP2C phosphatase | | Authors: | Dmitriev, A, Lovell, S, Battaile, K.P, Soules, K, Hefty, P.S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and ligand binding analyses of the periplasmic sensor domain of RsbU in Chlamydia trachomatis support a role in TCA cycle regulation.
Mol.Microbiol., 113, 2020
|
|
6VGY
 
 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor 6b | | Descriptor: | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH3
 
 | | 2.20 A resolution structure of MERS 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH2
 
 | | 2.26 A resolution structure of MERS 3CL protease in complex with inhibitor 7i | | Descriptor: | 4,4-difluorocyclohexyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH0
 
 | | 1.95 A resolution structure of MERS 3CL protease in complex with inhibitor 6g | | Descriptor: | N~2~-{[(5-ethyl-1,3-dioxan-5-yl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6VH1
 
 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
