1GXF
 
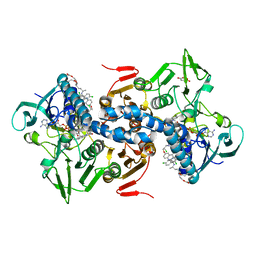 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH THE INHIBITOR QUINACRINE MUSTARD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALEIC ACID, QUINACRINE MUSTARD, ... | | Authors: | Bond, C.S, Peterson, M.R, Vickers, T.J, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2002-04-04 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two Interacting Binding Sites for Quinacrine Derivatives in the Active Site of Trypanothione Reductase: A Template for Drug Design
J.Biol.Chem., 279, 2004
|
|
7QJS
 
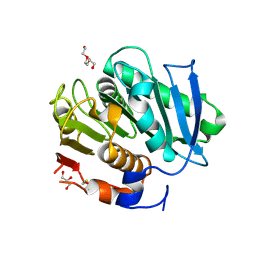 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
8GCB
 
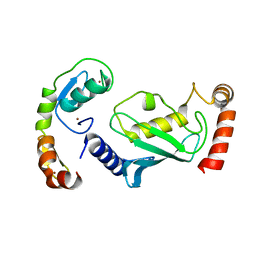 | | Structure of RNF125 in complex with a UbcH5b~Ub conjugate | | Descriptor: | E3 ubiquitin-protein ligase RNF125, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
8GBQ
 
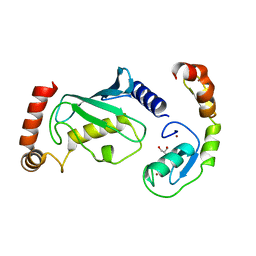 | | Structure of RNF125 in complex with UbcH5b | | Descriptor: | E3 ubiquitin-protein ligase RNF125, GLYCEROL, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Middleton, A.J, Day, C.L, Fokkens, T.J. | | Deposit date: | 2023-02-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Zinc finger 1 of the RING E3 ligase, RNF125, interacts with the E2 to enhance ubiquitylation.
Structure, 31, 2023
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
1HWY
 
 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | Deposit date: | 2001-01-10 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
5YI6
 
 | | CRISPR associated protein Cas6 | | Descriptor: | CRISPR-associated endoribonuclease Cas6 1, GLYCEROL, PHOSPHATE ION | | Authors: | Ko, T.P, Hsieh, T.J, Chen, Y. | | Deposit date: | 2017-10-03 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii
Crystals, 7, 2018
|
|
1HNA
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HRD
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
7RED
 
 | | Holo Hemophilin from A. baumannii | | Descriptor: | Hemophilin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
7RE4
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
7REA
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
8GY5
 
 | |
1VIH
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1D7A
 
 | | CRYSTAL STRUCTURE OF E. COLI PURE-MONONUCLEOTIDE COMPLEX. | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, PHOSPHORIBOSYLAMINOIMIDAZOLE CARBOXYLASE | | Authors: | Mathews, I.I, Kappock, T.J, Stubbe, J, Ealick, S.E. | | Deposit date: | 1999-10-16 | | Release date: | 1999-12-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway.
Structure Fold.Des., 7, 1999
|
|
1VIG
 
 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
5ZLR
 
 | | Structure of NeuC | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, SULFATE ION | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
5ZLT
 
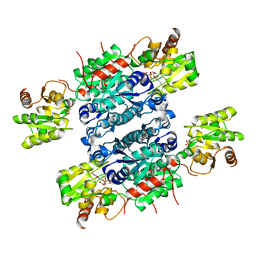 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Ko, T.P, Hsieh, T.J, Yang, C.S, Chen, Y. | | Deposit date: | 2018-03-29 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
8GI4
 
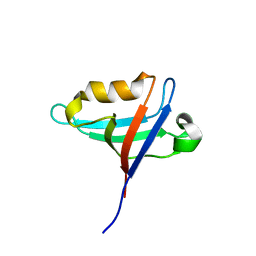 | |
8IQ9
 
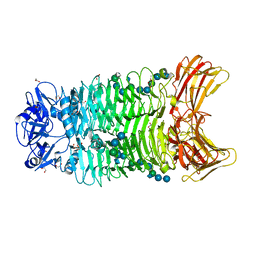 | | Crystal structure of trimeric K2-2 TSP in complex with tetrasaccharide and octasaccharide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, K2-2 TSP, ... | | Authors: | Ye, T.J, Ko, T.P, Huang, K.F, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
8IQE
 
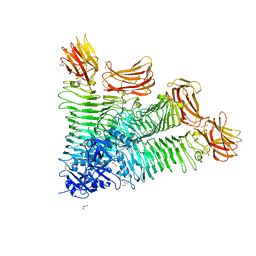 | | Crystal structure of tetrameric K2-2 TSP | | Descriptor: | GLYCEROL, K2-VCL6 TSP | | Authors: | Ye, T.J, Huang, K.F, Tu, I.F, Lee, I.M, Chang, Y.P, Wu, S.H. | | Deposit date: | 2023-03-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Klebsiella pneumoniae K2 capsular polysaccharide degradation by a bacteriophage depolymerase does not require trimer formation.
Mbio, 15, 2024
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
1PMB
 
 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
5XGU
 
 | | Escherichia coli. RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Chu, L.Y, Hsieh, T.J, Yuan, H.S. | | Deposit date: | 2017-04-17 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Structural insights into RNA unwinding and degradation by RNase R.
Nucleic Acids Res., 45, 2017
|
|
1PPA
 
 | | THE CRYSTAL STRUCTURE OF A LYSINE 49 PHOSPHOLIPASE A2 FROM THE VENOM OF THE COTTONMOUTH SNAKE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ANILINE, PHOSPHOLIPASE A2 | | Authors: | Holland, D.R, Clancy, L.L, Muchmore, S.W, Rydel, T.J, Einspahr, H.M, Finzel, B.C, Heinrikson, R.L, Watenpaugh, K.D. | | Deposit date: | 1991-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a lysine 49 phospholipase A2 from the venom of the cottonmouth snake at 2.0-A resolution.
J.Biol.Chem., 265, 1990
|
|
