7F62
 
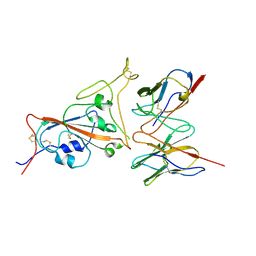 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region) | | Descriptor: | RBD-chAb-25, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
4EIJ
 
 | |
6U9U
 
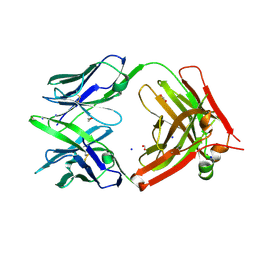 | | Structure of GM9_TH8seq732127 FAB | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GM9_TH8seq732127 FAB heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
4EPA
 
 | |
3KXX
 
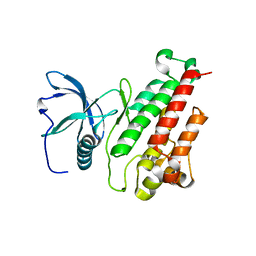 | | Structure of the mutant Fibroblast Growth Factor receptor 1 | | Descriptor: | Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Boggon, T.J, Tome, F, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4WAS
 
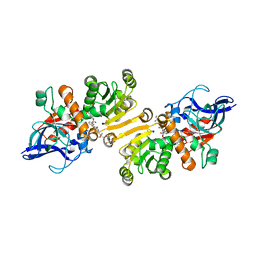 | | STRUCTURE OF THE ETR1P/NADP/CROTONYL-COA COMPLEX | | Descriptor: | CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Capitani, G, Erb, T.J. | | Deposit date: | 2014-08-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
4EAH
 
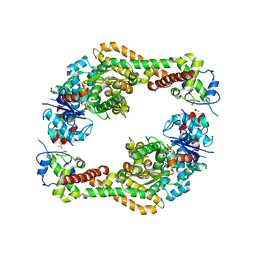 | | Crystal structure of the formin homology 2 domain of FMNL3 bound to actin | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Thompson, M.E, Heimsath, E.G, Gauvin, T.J, Higgs, H.N, Kull, F.J. | | Deposit date: | 2012-03-22 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4EDM
 
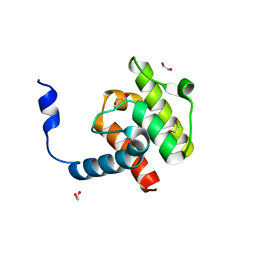 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
7R6R
 
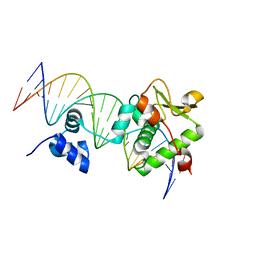 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
8QMW
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitutions R269W, E271R, L273N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Layered entrenchment maintains essentiality in protein-protein interactions
Embo J., 2024
|
|
4F3V
 
 | |
8QMQ
 
 | | Succinic semialdehyde dehydrogenase from E. coli with bound NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMV
 
 | | L2 forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Layered entrenchment maintains essentiality in the evolution of Form I Rubisco complexes
Biorxiv, 2024
|
|
8QMR
 
 | |
8QMT
 
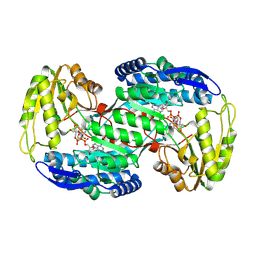 | | Succinic semialdehyde dehydrogenase from E. coli with Q262R substitution and bound NAD+, succinic semialdehyde | | Descriptor: | 4-oxobutanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Succinate semialdehyde dehydrogenase [NAD(P)+] Sad | | Authors: | He, H, Zarzycki, J, Erb, T.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Adaptive laboratory evolution recruits the promiscuity of succinate semialdehyde dehydrogenase to repair different metabolic deficiencies.
Nat Commun, 15, 2024
|
|
8QMS
 
 | |
7RIZ
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound 2-hydroxyquinoline | | Descriptor: | Beta-propeller lactonase, CALCIUM ION, QUINOLIN-2(1H)-ONE, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2021-07-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
7RIS
 
 | | Crystal structure of RPA3624, a beta-propeller lactonase from Rhodopseudomonas palustris, with active-site bound phosphate | | Descriptor: | Beta-propeller lactonase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Bingman, C.A, Hall, B.W, Smith, R.W, Fox, B.G, Donohue, T.J. | | Deposit date: | 2021-07-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A broad specificity beta-propeller enzyme from Rhodopseudomonas palustris that hydrolyzes many lactones including gamma-valerolactone.
J.Biol.Chem., 299, 2022
|
|
4WJ7
 
 | | CCM2 PTB domain in complex with KRIT1 NPxY/F3 | | Descriptor: | KRIT1 NPxY/F3, Malcavernin | | Authors: | Fisher, O.S, Liu, W, Zhang, R, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structural Basis for the Disruption of the Cerebral Cavernous Malformations 2 (CCM2) Interaction with Krev Interaction Trapped 1 (KRIT1) by Disease-associated Mutations.
J.Biol.Chem., 290, 2015
|
|
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3L8I
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
6VE6
 
 | | A structural characterization of poly(aspartic acid) hydrolase-1 from Sphingomonas sp. KT-1. | | Descriptor: | Poly(Aspartic acid) hydrolase-1 | | Authors: | Bolay, A.L, Salvo, H, Brambley, C.A, Yared, T.J, Miller, J.M, Wallen, J.R, Weiland, M.H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Structural Characterization of Sphingomonas sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
Acs Sustain Chem Eng, 2020
|
|
6N95
 
 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-CoA | | Descriptor: | (2R)-sulfonatepropionyl-CoA, (2S)-sulfonatepropionyl-CoA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
7SGY
 
 | |
7SHU
 
 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
