1ZAL
 
 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
4KOO
 
 | | Crystal Structure of WHY1 from Arabidopsis thaliana | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOP
 
 | | Crystal Structure of WHY2 from Arabidopsis thaliana | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Single-stranded DNA-binding protein WHY2, mitochondrial | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4KOQ
 
 | | Crystal Structure of WHY3 from Arabidopsis thaliana | | Descriptor: | PHOSPHATE ION, Single-stranded DNA-binding protein WHY3, chloroplastic | | Authors: | Cappadocia, L, Parent, J.S, Brisson, N, Sygusch, J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ZR5
 
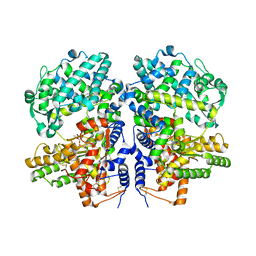 | | Soluble rabbit neprilysin in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2015-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8016 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3RA0
 
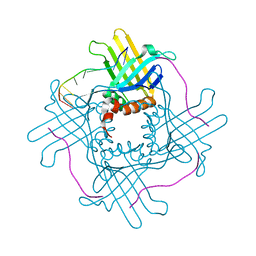 | |
3R9Y
 
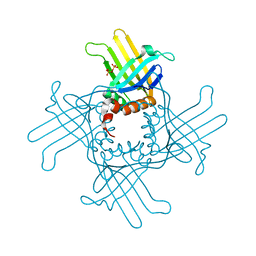 | | Crystal Structure of StWhy2 K67A (form I) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
3R9Z
 
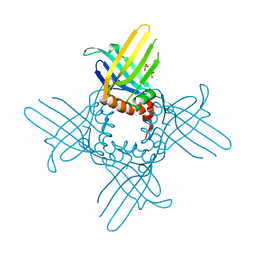 | | Crystal Structure of StWhy2 K67A (form II) | | Descriptor: | PHOSPHATE ION, Why2 protein | | Authors: | Cappadocia, L, Brisson, N, Sygusch, J. | | Deposit date: | 2011-03-26 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage.
Nucleic Acids Res., 40, 2012
|
|
2QAP
 
 | |
2QDH
 
 | | Fructose-1,6-bisphosphate aldolase from Leishmania mexicana in complex with mannitol-1,6-bisphosphate, a competitive inhibitor | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, Fructose-1,6-bisphosphate aldolase | | Authors: | Lafrance-Vanasse, J, Sygusch, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carboxy-Terminus Recruitment Induced by Substrate Binding in Eukaryotic Fructose Bis-phosphate Aldolases
Biochemistry, 46, 2007
|
|
2QDG
 
 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from Leishmania mexicana | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-1,6-bisphosphate aldolase, PHOSPHATE ION | | Authors: | Lafrance-Vanasse, J, Sygusch, J. | | Deposit date: | 2007-06-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxy-Terminus Recruitment Induced by Substrate Binding in Eukaryotic Fructose Bis-phosphate Aldolases
Biochemistry, 46, 2007
|
|
1ADO
 
 | | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE FROM RABBIT MUSCLE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, ALDOLASE, SULFATE ION | | Authors: | Blom, N.S, Sygusch, J. | | Deposit date: | 1996-12-02 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 4, 1997
|
|
1DOS
 
 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | Descriptor: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | Authors: | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | Deposit date: | 1996-06-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
4A22
 
 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to N-(4-hydroxybutyl)- glycolohydroxamic acid bis- phosphate | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, ... | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli.
J. Biol. Chem., 286, 2011
|
|
1EUN
 
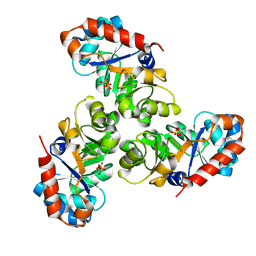 | |
1EUA
 
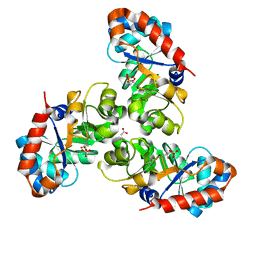 | | SCHIFF BASE INTERMEDIATE IN KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | ACETATE ION, KDPG ALDOLASE, PYRUVIC ACID, ... | | Authors: | Allard, J, Grochulski, P, Sygusch, J. | | Deposit date: | 2000-04-14 | | Release date: | 2001-02-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2OT0
 
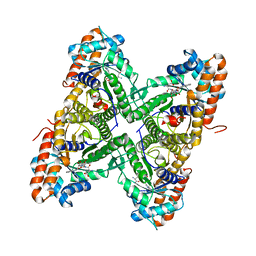 | |
2OT1
 
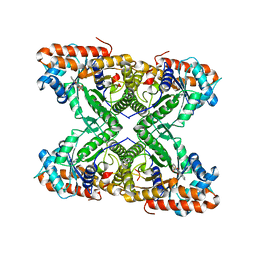 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with naphthol AS-E phosphate, a competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, N-(4-CHLOROPHENYL)-3-(PHOSPHONOOXY)NAPHTHALENE-2-CARBOXAMIDE | | Authors: | St-Jean, M, Izard, T, Sygusch, J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with wiskott-Aldrich syndrome protein.
J.Biol.Chem., 282, 2007
|
|
2QUV
 
 | |
2QUU
 
 | |
2QUT
 
 | |
3LGE
 
 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
3N1I
 
 | |
3N1J
 
 | |
3N1L
 
 | |
