2I3O
 
 | |
2GV8
 
 | | Crystal structure of flavin-containing monooxygenase (FMO) from S.pombe and NADPH cofactor complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HZT
 
 | | Crystal Structure of a putative HTH-type transcriptional regulator ytcD | | Descriptor: | Putative HTH-type transcriptional regulator ytcD | | Authors: | Madegowda, M, Eswaramoorthy, S, Desigan, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-29 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a putative HTH-type transcription regulator ytcD
To be Published
|
|
2I5H
 
 | |
1YXW
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | GLUTAMIC ACID, TRYPTOPHAN, TYROSINE, ... | | Authors: | Jayaraman, S, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
1YYN
 
 | | A common binding site for disialyllactose and a tri-peptide in the C-fragment of tetanus neurotoxin | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-alpha-D-galactopyranose-(1-4)-beta-D-glucopyranose, Tetanus toxin | | Authors: | Seetharaman, J, Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common binding site for disialyllactose and tri-peptide in C-fragment of tetanus neurotoxin
Proteins, 61, 2005
|
|
1Z0H
 
 | | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Jayaraman, S, Eswarmoorthy, S, Ashraf, S.A, Smith, L.A, Swaminathan, S. | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
2ICS
 
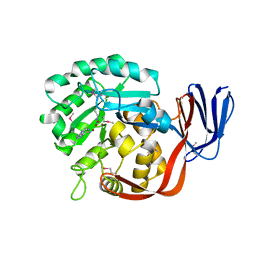 | | Crystal structure of an adenine deaminase | | Descriptor: | ADENINE, Adenine Deaminase, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine deaminase
TO BE PUBLISHED
|
|
2IMO
 
 | |
2IMG
 
 | | Crystal structure of dual specificity protein phosphatase 23 from Homo sapiens in complex with ligand malate ion | | Descriptor: | D-MALATE, Dual specificity protein phosphatase 23 | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex.
J.Biol.Chem., 283, 2008
|
|
2IOJ
 
 | |
2HAE
 
 | |
2I6E
 
 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
2I9U
 
 | |
2ABQ
 
 | |
2QJC
 
 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2NN4
 
 | | Crystal structure of Bacillus subtilis yqgQ, Pfam DUF910 | | Descriptor: | Hypothetical protein yqgQ | | Authors: | Damodharan, L, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of YqgQ protein from Bacillus subtilis, a conserved hypothetical protein.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2OOF
 
 | |
2PB9
 
 | |
2NYG
 
 | |
2POF
 
 | |
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
2PHP
 
 | |
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2QQ6
 
 | |
