5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
5YC8
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS (Hg-derivative) | | Descriptor: | MERCURY (II) ION, Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZK8
 
 | | Crystal structure of M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Redesigned apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKB
 
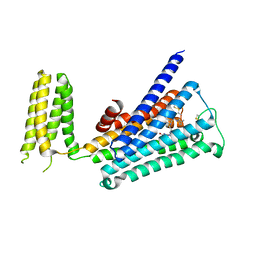 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
5WS3
 
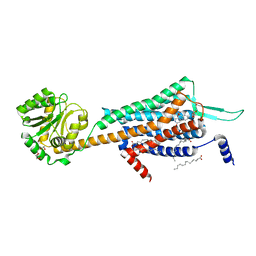 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
5ZK3
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with QNB | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2 | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
3SBJ
 
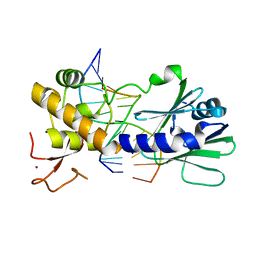 | | MutM slanted complex 7 | | Descriptor: | 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*G)-3', 5'-D(P*CP*CP*TP*GP*GP*TP*(CX)P*TP*AP*CP*C)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Verdine, G.L. | | Deposit date: | 2011-06-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strandwise translocation of a DNA glycosylase on undamaged DNA.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U6Q
 
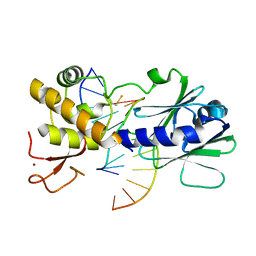 | | MutM set 2 ApGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6L
 
 | | MutM set 2 CpGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2017-01-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6M
 
 | | Structural effects of sequence context on lesion recognition by MutM | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*AP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*TP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
2DHR
 
 | | Whole cytosolic region of ATP-dependent metalloprotease FtsH (G399L) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FtsH | | Authors: | Suno, R, Niwa, H, Tsuchiya, D, Zhang, X, Yoshida, M, Morikawa, K. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH
Mol.Cell, 22, 2006
|
|
3U6P
 
 | | MutM set 1 GpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6O
 
 | | MutM set 1 ApG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*TP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*AP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6C
 
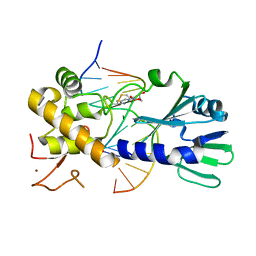 | | MutM set 1 ApGo | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*GP*AP*TP*CP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*CP*AP*(8OG)P*GP*AP*(TX)P*CP*TP*AP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6D
 
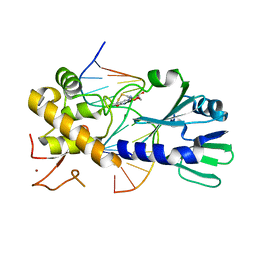 | | MutM set 1 GpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*(8OG)P*GP*AP*(TX)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6E
 
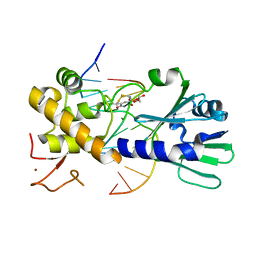 | | MutM set 1 TpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*(8OG)P*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6S
 
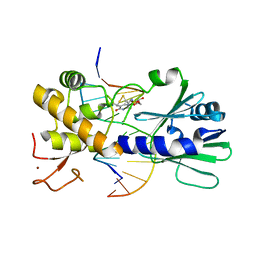 | | MutM set 1 TpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
5M00
 
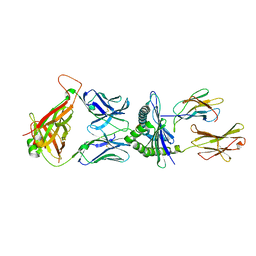 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
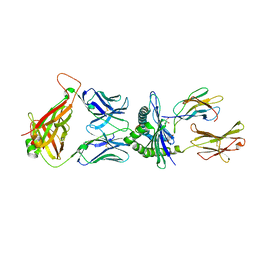 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M02
 
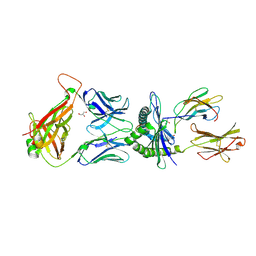 | |
6EWQ
 
 | | Putative sugar aminotransferase Spr1654 from Streptococcus pneumoniae, PLP-form | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative capsular polysaccharide biosynthesis protein | | Authors: | Achour, A, Sun, R, Sandalova, T, Han, X. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional studies of Spr1654: an essential aminotransferase in teichoic acid biosynthesis inStreptococcus pneumoniae.
Open Biol, 8, 2018
|
|
6EWJ
 
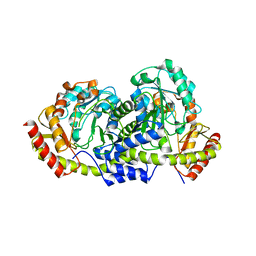 | | Putative sugar aminotransferase Spr1654 from Streptococcus pneumoniae, apo-form | | Descriptor: | Putative capsular polysaccharide biosynthesis protein | | Authors: | Achour, A, Sun, R, Sandalova, T, Han, X. | | Deposit date: | 2017-11-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of Spr1654: an essential aminotransferase in teichoic acid biosynthesis inStreptococcus pneumoniae.
Open Biol, 8, 2018
|
|
6EWR
 
 | | Putative sugar aminotransferase Spr1654 from Streptococcus pneumoniae, PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Putative capsular polysaccharide biosynthesis protein | | Authors: | Achour, A, Sun, R, Sandalova, T, Han, X. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional studies of Spr1654: an essential aminotransferase in teichoic acid biosynthesis inStreptococcus pneumoniae.
Open Biol, 8, 2018
|
|
6G7C
 
 | | Nt2-CTD domains of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | Descriptor: | ImpA-related domain protein | | Authors: | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-04-05 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
