8HO8
 
 | |
8HOB
 
 | |
6N8Z
 
 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2019-01-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N8T
 
 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6NZ6
 
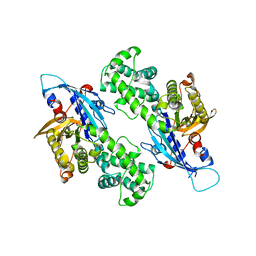 | | YcjX-GDP (type II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Tsai, J, Sung, N, Lee, J, Chang, C, Lee, S, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6NZ5
 
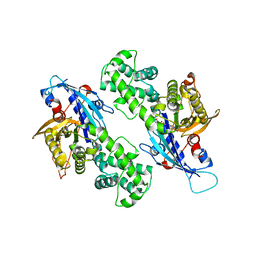 | | YcjX-GDPCP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T, Sung, N, Lee, J, Chang, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
5V2W
 
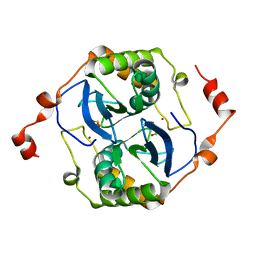 | | Crystal structure of a LuxS from salmonella typhi | | Descriptor: | S-ribosylhomocysteine lyase, ZINC ION | | Authors: | Perumal, P, Raina, R, Manoj Kumar, P, Arockisamy, A, SundaraBaalaji, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a LuxS from salmonella typhi
To Be Published
|
|
7VC7
 
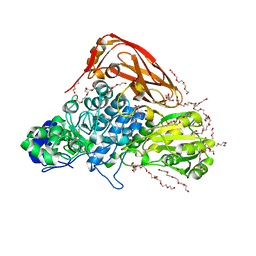 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
7VC6
 
 | | The structure of beta-xylosidase from Phanerochaete chrysosporium(PcBxl3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, xylan 1,4-beta-xylosidase | | Authors: | Kojima, K, Sunagawa, N, Igarashi, K. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Comparison of glycoside hydrolase family 3 beta-xylosidases from basidiomycetes and ascomycetes reveals evolutionarily distinct xylan degradation systems.
J.Biol.Chem., 298, 2022
|
|
4FD2
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCT
 
 | |
4FCV
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCW
 
 | | Crystal structure of the C-terminal domain of ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Biter, A.B, Lee, S, Sung, N, Tsai, F.T.F. | | Deposit date: | 2012-05-25 | | Release date: | 2012-07-18 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for intersubunit signaling in a protein disaggregating machine.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7EZZ
 
 | | Crystal structure of Salmonella typhi outer membrane phospholipase (OMPLA) dimer with bound calcium | | Descriptor: | CALCIUM ION, DODECANE, Phospholipase A1, ... | | Authors: | Perumal, P, Raina, R, Sreeshma, N.S, Arockiasamy, A, Sundarabaalaji, N. | | Deposit date: | 2021-06-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Salmonella typhi outer membrane phospholipase (OMPLA) dimer with bound calcium
To Be Published
|
|
