5Y6P
 
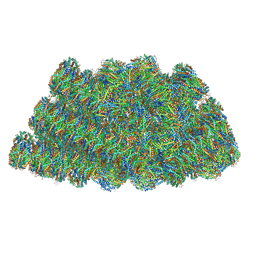 | | Structure of the phycobilisome from the red alga Griffithsia pacifica | | 分子名称: | ApcD, ApcF, LC, ... | | 著者 | Zhang, J, Ma, J.F, Liu, D.S, Sun, S, Sui, S.F. | | 登録日 | 2017-08-13 | | 公開日 | 2017-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of phycobilisome from the red alga Griffithsia pacifica
Nature, 551, 2017
|
|
8JNN
 
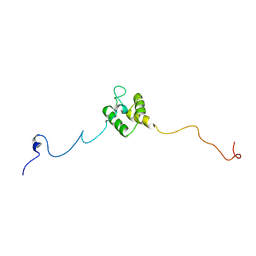 | | linker protein LPP1 from red algal Porphyridium purpureum. | | 分子名称: | LPP1 | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNG
 
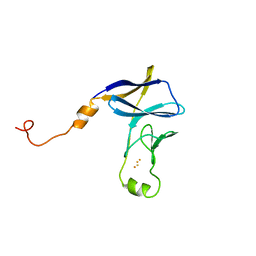 | | Methanesulfonate monooxygenase ferredoxin subunit of PBS-PSII-PSI-LHCs from Porphyridium purpureum. | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Methanesulfonate monooxygenase ferredoxin subunit | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
7WOT
 
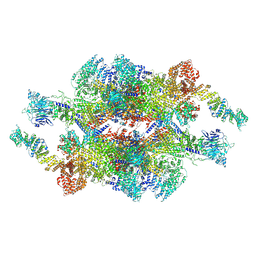 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | 分子名称: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | 著者 | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | 登録日 | 2022-01-22 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7K4Y
 
 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
8JNL
 
 | | Psb34 from red algal Porphyridium purpureum. | | 分子名称: | Psb34 | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
8JNM
 
 | | PsbW from red algal Porphyridium purpureum. | | 分子名称: | PsbW | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of lateral hexamer of PBS-PSII-PSI-LHCs megacomplex at 6.3 Angstroms resolution.
To Be Published
|
|
7VD2
 
 | | Human TOM complex without cross-linking | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | 著者 | Liu, D.S, Sui, S.F. | | 登録日 | 2021-09-06 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WOO
 
 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | 分子名称: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | 著者 | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | 登録日 | 2022-01-22 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7VDD
 
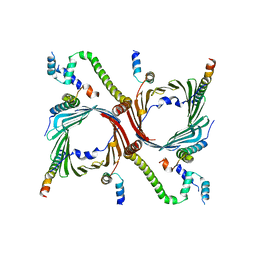 | | Human TOM complex with cross-linking | | 分子名称: | Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | 著者 | Liu, D.S, Sui, S.F. | | 登録日 | 2021-09-06 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WO9
 
 | |
7K4S
 
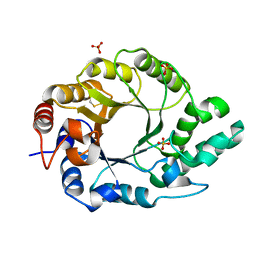 | | Crystal structure of Kemp Eliminase HG3.7 | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4X
 
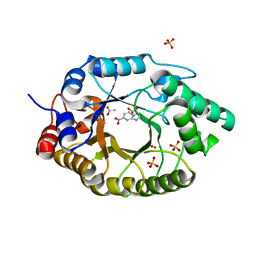 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Q
 
 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4R
 
 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4P
 
 | | Crystal structure of Kemp Eliminase HG3 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
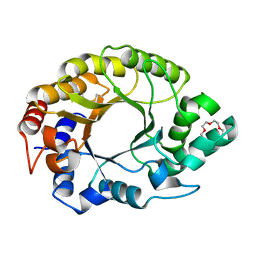 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4W
 
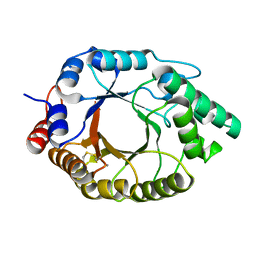 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4U
 
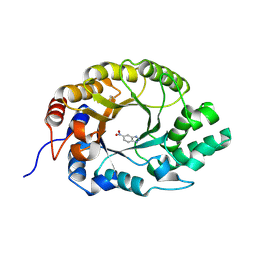 | | Crystal structure of Kemp Eliminase HG3 K50Q in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7VZR
 
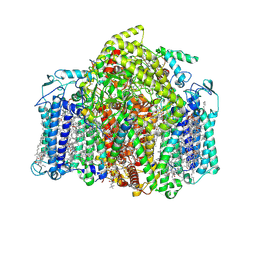 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | 著者 | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | 登録日 | 2021-11-16 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.22 Å) | | 主引用文献 | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7VZG
 
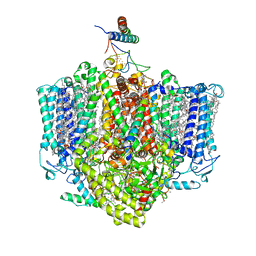 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | 著者 | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | 登録日 | 2021-11-16 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
7VBY
 
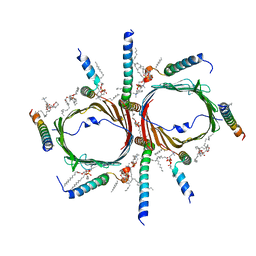 | | Tom core complex with Tom20 and Tom22 subunits. | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | 著者 | Liu, D.S, Sui, S.F. | | 登録日 | 2021-09-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.54 Å) | | 主引用文献 | Tom core complex with Tom20 and Tom22 subunits.
To Be Published
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | 分子名称: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
