3AWZ
 
 | |
3AWU
 
 | |
3AWX
 
 | |
2ZMX
 
 | |
3WSW
 
 | |
3WSV
 
 | |
3VVM
 
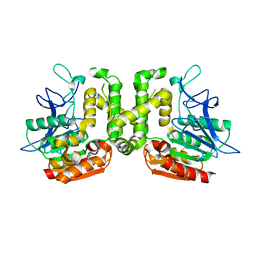 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | Descriptor: | Homoserine O-acetyltransferase | | Authors: | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3AZQ
 
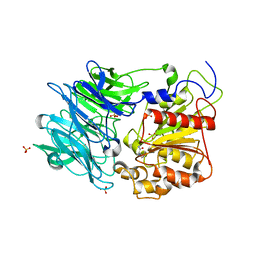 | |
3AZO
 
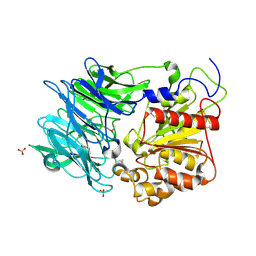 | | Crystal structure of puromycin hydrolase | | Descriptor: | Aminopeptidase, SULFATE ION | | Authors: | Matoba, Y, Sugiyama, M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that puromycin hydrolase is a new type of aminopeptidase with a prolyl oligopeptidase family fold
Proteins, 79, 2011
|
|
3AZP
 
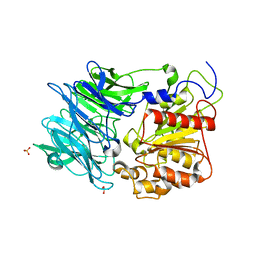 | |
3WA6
 
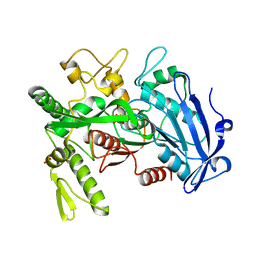 | |
3WA7
 
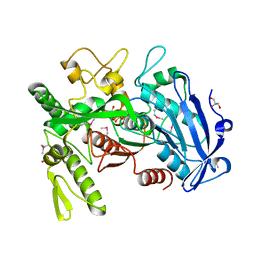 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Matoba, Y, Tanaka, N, Sugiyama, M. | | Deposit date: | 2013-04-27 | | Release date: | 2013-07-24 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
2ZRR
 
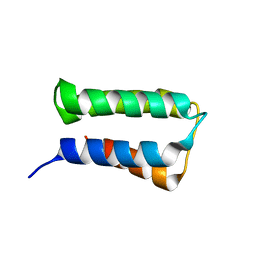 | | Crystal structure of an immunity protein that contributes to the self-protection of bacteriocin-producing Enterococcus mundtii 15-1A | | Descriptor: | Mundticin KS immunity protein | | Authors: | Jeon, H.J, Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2008-08-30 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im
Biochem.Biophys.Res.Commun., 378, 2009
|
|
2ZW4
 
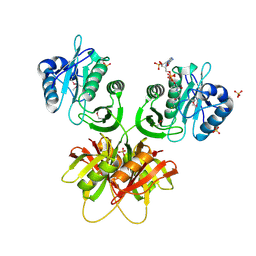 | |
2ZW7
 
 | |
2ZW5
 
 | |
2ZW6
 
 | |
3X44
 
 | | Crystal structure of O-ureido-L-serine-bound K43A mutant of O-ureido-L-serine synthase | | Descriptor: | (E)-O-(carbamoylamino)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, O-ureido-L-serine synthase | | Authors: | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
3X43
 
 | | Crystal structure of O-ureido-L-serine synthase | | Descriptor: | O-ureido-L-serine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
2ZV6
 
 | | Crystal structure of human squamous cell carcinoma antigen 1 | | Descriptor: | Serpin B3 | | Authors: | Zheng, B, Matoba, Y, Katagiri, C, Hibino, T, Sugiyama, M. | | Deposit date: | 2008-11-01 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SCCA1 and insight about the interaction with JNK1
Biochem.Biophys.Res.Commun., 380, 2009
|
|
