3L7O
 
 | |
3L7Q
 
 | |
3L7N
 
 | |
3URG
 
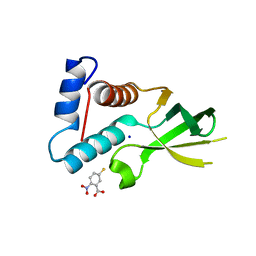 | | The crystal structure of Anabaena CcbP | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Alr1010 protein, SODIUM ION | | Authors: | Fan, X.X, Liu, X, Su, X.D. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Ellman's reagent in promoting crystallization and structure determination of Anabaena CcbP.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5EMQ
 
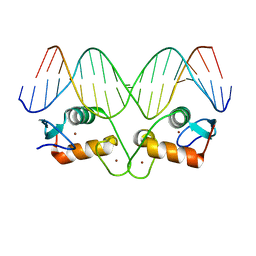 | | Transcription factor GRDBD and GRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*AP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*AP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Lian, T, Jin, J, Su, X. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effects of cytosine methylation on general transcription factors
To Be Published
|
|
3E4Q
 
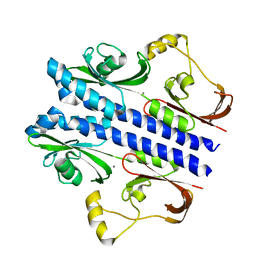 | | Crystal structure of apo DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, CALCIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
5EMP
 
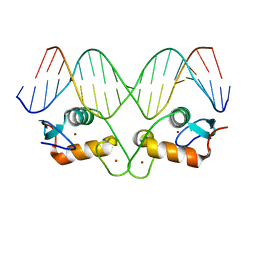 | | Transcription factor GRDBD and mmGRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*AP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*CP*AP*GP*AP*AP*CP*AP*TP*(5CM)P*AP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Lian, T, Jin, J, Su, X. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effects of cytosine methylation on general transcription factors
To Be Published
|
|
4L22
 
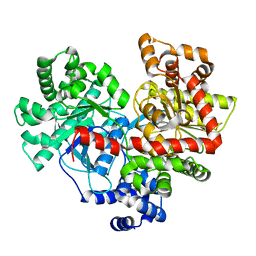 | |
3F7J
 
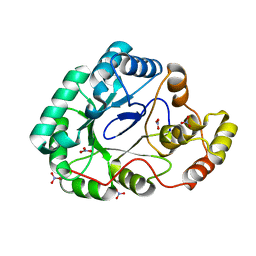 | | B.subtilis YvgN | | Descriptor: | NITRATE ION, POTASSIUM ION, YvgN protein | | Authors: | Zhou, Y.F, Lei, J, Liang, Y.H, Su, X.-D. | | Deposit date: | 2008-11-09 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3DEZ
 
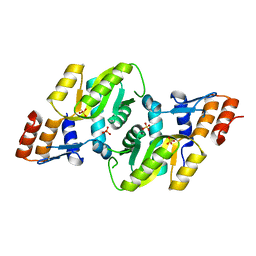 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | Descriptor: | Orotate phosphoribosyltransferase, SULFATE ION | | Authors: | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3E4P
 
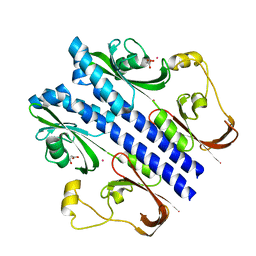 | | Crystal structure of malonate occupied DctB | | Descriptor: | C4-dicarboxylate transport sensor protein dctB, MALONIC ACID, STRONTIUM ION | | Authors: | Zhou, Y.F, Nan, J, Nan, B.Y, Liang, Y.H, Panjikar, S, Su, X.D. | | Deposit date: | 2008-08-12 | | Release date: | 2008-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain.
J.Mol.Biol., 383, 2008
|
|
2K7R
 
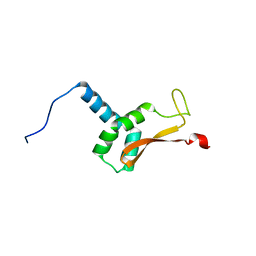 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
3LCM
 
 | | Crystal structure of Smu.1420 from Streptococcus mutans UA159 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Wang, Z.X, Su, X.-D. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural and biochemical characterization of MdaB from cariogenic Streptococcus mutans reveals an NADPH-specific quinone oxidoreductase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3L8U
 
 | |
3L78
 
 | |
3LAS
 
 | | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution | | Descriptor: | GLYCEROL, GUANIDINE, MAGNESIUM ION, ... | | Authors: | Ma, L.-L, Wang, K.-T, Liu, X, Su, X.-D. | | Deposit date: | 2010-01-07 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution
To be published
|
|
3SR7
 
 | |
3IJT
 
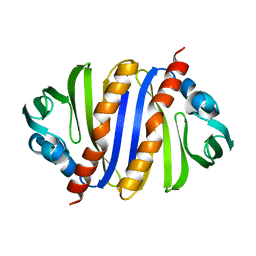 | | Structural Characterization of SMU.440, a Hypothetical Protein from Streptococcus mutans | | Descriptor: | Putative uncharacterized protein | | Authors: | Nan, J, Brostromer, E, Kristensen, O, Su, X.-D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance
Plos One, 4, 2009
|
|
3SQZ
 
 | |
3LE5
 
 | |
3V6L
 
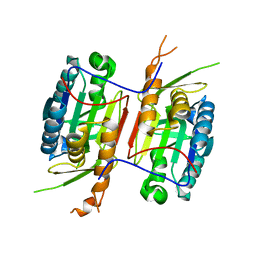 | | Crystal Structure of caspase-6 inactivation mutation | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
3V6M
 
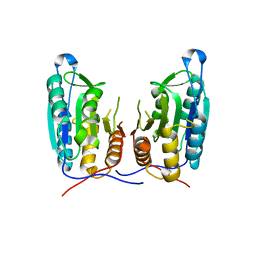 | | Inhibition of caspase-6 activity by single mutation outside the active site | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.J, Liu, D.F, Li, L.F, Su, X.D. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays
J.Biol.Chem., 287, 2012
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6IJO
 
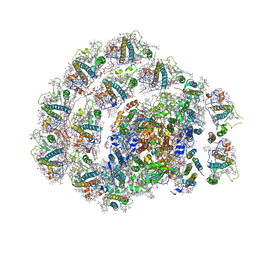 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
3L7R
 
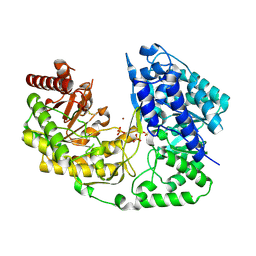 | | crystal structure of MetE from streptococcus mutans | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, SULFATE ION, ZINC ION | | Authors: | Fu, T.M, Liang, Y.H, Su, X.D. | | Deposit date: | 2009-12-29 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal Structures of Cobalamin-Independent Methionine Synthase (MetE) from Streptococcus mutans: A Dynamic Zinc-Inversion Model
J.Mol.Biol., 412, 2011
|
|
