5GZF
 
 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
7FG9
 
 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
4RCB
 
 | | Crystal structure of E Coli Hfq | | Descriptor: | RNA-binding protein Hfq | | Authors: | Su, J.Y. | | Deposit date: | 2014-09-15 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Limited proteolysis improves E.coli Hfq crystal structure resolution
Chin J Biochem Mol Biol, 9, 2015
|
|
7VSS
 
 | | Phosphoglucomutase_tlr1976 | | Descriptor: | Phosphoglucomutase | | Authors: | Su, J.Y. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | phosphoglucomutase_tlr1976
To Be Published
|
|
7VST
 
 | | Phosphoglucomutase_tlr1976 | | Descriptor: | Phosphoglucomutase | | Authors: | Su, J.Y. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Phosphoglucomutase_tlr1976
To Be Published
|
|
5ITI
 
 | | A cynobacterial PP2C (tPphA) structure | | Descriptor: | CALCIUM ION, Protein serin-threonin phosphatase | | Authors: | Su, J.Y. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of a Cyanobacterial PP2C Phosphatase Reveals Insights into Catalytic Mechanism and Substrate Recognition
Catalysts, 6, 2016
|
|
8XRT
 
 | | The crystal structure of a GH3 enzyme CcBgl3B | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRV
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRX
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose and gentiobiose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, alpha-D-glucopyranose, ... | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRU
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glycerol | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, GLYCEROL | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8X3Q
 
 | | tll1591 with alpha-glucan 4sugar | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2023-11-14 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Glucosyltransferase TeGSS from Thermosynechococcus elongatus produces an alpha-1,2-glucan.
Int.J.Biol.Macromol., 280, 2024
|
|
8X3U
 
 | | tll1591 with alpha_glucan 3sugar | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2023-11-14 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Glucosyltransferase TeGSS from Thermosynechococcus elongatus produces an alpha-1,2-glucan.
Int.J.Biol.Macromol., 280, 2024
|
|
8WXK
 
 | |
8X7S
 
 | | Crystal structure of tlr0611 | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Su, J.Y. | | Deposit date: | 2023-11-25 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of tlr0611
To Be Published
|
|
6A66
 
 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
5XG8
 
 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | GLYCEROL, Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J.Y. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138
Sci Rep, 8, 2018
|
|
8HWL
 
 | | Human Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (5.63 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 2024
|
|
8J4Z
 
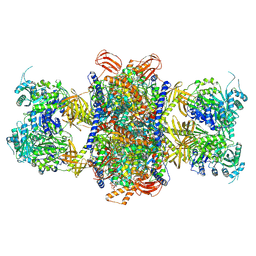 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-CTS state with substrate | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J7O
 
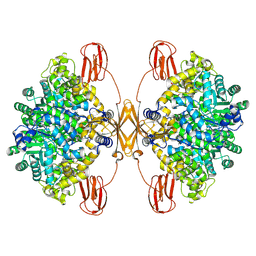 | | Human pyruvate carboxylase in BCCP-CTS state without BC | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J73
 
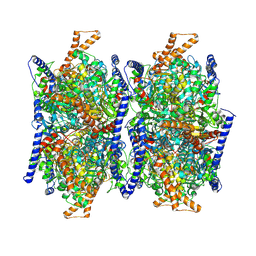 | | Methylcrotonoyl-CoA carboxylase core-Dimer | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J78
 
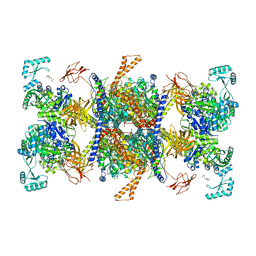 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8J7D
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JAK
 
 | | Human MCC in MCCU state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8JAW
 
 | | Human MCC in MCCD state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-07 | | Release date: | 2024-05-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
