2BMY
 
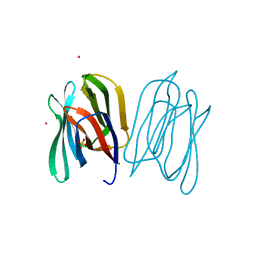 | | Banana Lectin | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
3PBJ
 
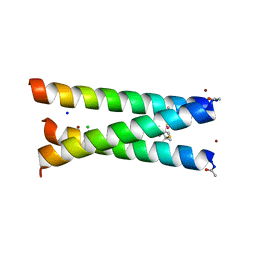 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | Descriptor: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | Authors: | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-10-20 | | Release date: | 2011-11-30 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
3LQ6
 
 | |
3LQE
 
 | |
1LW3
 
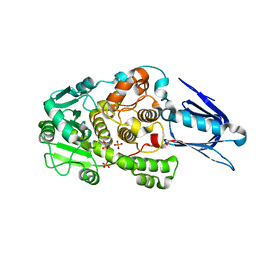 | | Crystal Structure of Myotubularin-related protein 2 complexed with phosphate | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-05-30 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Phosphoinositide Phosphatase, MTMR2: Insights into Myotubular Myopathy and Charcot-Marie-Tooth Syndrome
Mol.Cell, 12, 2003
|
|
1M7R
 
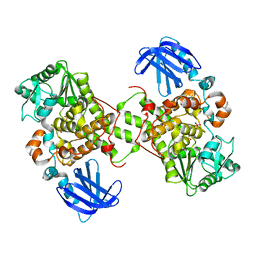 | | Crystal Structure of Myotubularin-related Protein-2 (MTMR2) Complexed with Phosphate | | Descriptor: | Myotubularin-related Protein-2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-07-22 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome
Mol.Cell, 12, 2003
|
|
5KB2
 
 | | Crystal Structure of a Tris-thiolate Zn(II)S3O Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | ZINC ION, Zn(II)(H2O)(GRAND Coil Ser-L12AL16C)3- | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5KB1
 
 | | Crystal Structure of a Tris-thiolate Hg(II) Complex in a de Novo Three Stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, Hg(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, MERCURY (II) ION, ... | | Authors: | Ruckcthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5K92
 
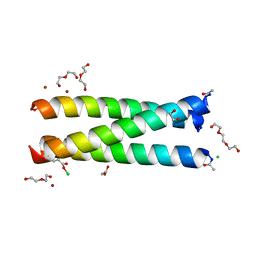 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
4J09
 
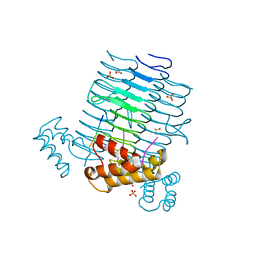 | | Crystal Structure of LpxA bound to RJPXD33 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Jenkins, R.J, Meagher, J.L, Stuckey, J.A, Dotson, G.D. | | Deposit date: | 2013-01-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
J.Biol.Chem., 289, 2014
|
|
5KB0
 
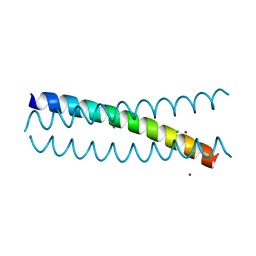 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
7N2Y
 
 | |
7N2Z
 
 | | Crystal Structure of a de Novo Three-stranded Coiled Coil Peptide Containing Trigonal Pyrmidal Pb(II) complexes in the dual Tris-thiolate Site | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)2-(GRAND CoilSerL16CL23C)3, ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Open Reading Frame 1 Protein of the Human Long Interspersed Nuclear Element 1 Retrotransposon Binds Multiple Equivalents of Lead.
J.Am.Chem.Soc., 143, 2021
|
|
3H5G
 
 | |
3H5F
 
 | | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides. | | Descriptor: | ACETATE ION, CHLORIDE ION, COIL SER L16L-Pen, ... | | Authors: | Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2009-04-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Switching the Chirality of the Metal Environment Alters the Coordination Mode in Designed Peptides.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6U9M
 
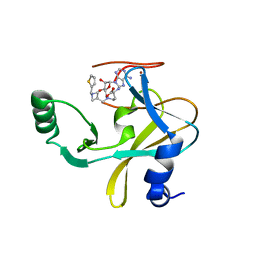 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6MCD
 
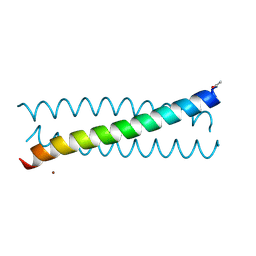 | | Crystal Structure of tris-thiolate Pb(II) complex with adjacent water in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | LEAD (II) ION, Pb(II)(GRAND Coil Ser L12CL16A)-, ZINC ION | | Authors: | Tolbert, A.E, Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-31 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteromeric three-stranded coiled coils designed using a Pb(II)(Cys)3template mediated strategy.
Nat.Chem., 12, 2020
|
|
6MHB
 
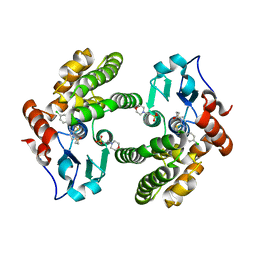 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MHD
 
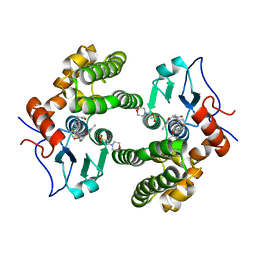 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 44 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETONE, Glutathione S-transferase omega-1, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MHC
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 37 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
1LRN
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LRO
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP and Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LRQ
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
