6VFX
 
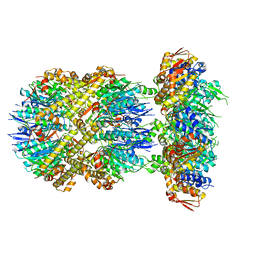 | | ClpXP from Neisseria meningitidis - Conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
6VGN
 
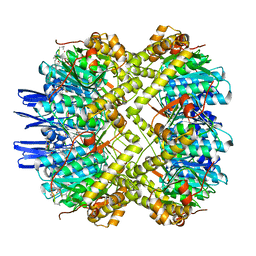 | | ClpP1P2 complex from M. tuberculosis bound to ADEP | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, R0M-WFP-ALO-PRO-YCP-ALA-MP8 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7UWA
 
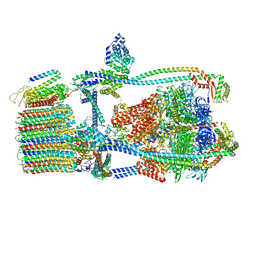 | | Citrus V-ATPase State 1, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Abdelaziz, R.A, Keon, K.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UW9
 
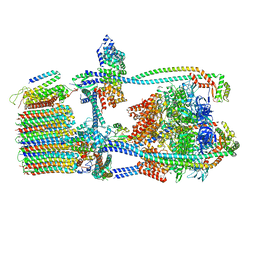 | | Citrus V-ATPase State 1, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWB
 
 | | Citrus V-ATPase State 2, Highest-Resolution Class | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWD
 
 | | Citrus V-ATPase State 2, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit, V-type proton ATPase subunit AP1 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UWC
 
 | | Citrus V-ATPase State 2, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7TKP
 
 | |
7TKI
 
 | |
7TKM
 
 | |
7TKO
 
 | |
7TKH
 
 | |
7TKN
 
 | |
7TKR
 
 | |
7TKF
 
 | |
7TKK
 
 | |
7TKG
 
 | |
7TKQ
 
 | |
7TKJ
 
 | |
7TKS
 
 | |
7TKL
 
 | |
7UZK
 
 | |
7UZJ
 
 | |
6VFS
 
 | | ClpXP from Neisseria meningitidis - Conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
1MXS
 
 | | Crystal structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. | | Descriptor: | KDPG Aldolase, SULFATE ION | | Authors: | Watanabe, L, Bell, B.J, Lebioda, L, Rios-Steiner, J.L, Tulinsky, A, Arni, R.K. | | Deposit date: | 2002-10-03 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
