4Q4X
 
 | |
3S6X
 
 | |
6Y5X
 
 | |
6Y6A
 
 | | Structure of Finch Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | CHLORIDE ION, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
6Y66
 
 | | Structure of Goose Hemorrhagic Polyomavirus VP1 in complex with 2-O-Methyl-5-N-acetyl-alpha-D-neuraminic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, Capsid protein VP1, ... | | Authors: | Stroh, L.J, Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis and Evolution of Glycan Receptor Specificities within the Polyomavirus Family.
Mbio, 11, 2020
|
|
3S6Z
 
 | |
4Q4Y
 
 | |
3TW5
 
 | | Crystal structure of the GP42 transglutaminase from Phytophthora sojae | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Transglutaminase elicitor | | Authors: | Reiss, K, Kirchner, E, Zocher, G, Stehle, T. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and Phylogenetic Analyses of the GP42 Transglutaminase from Phytophthora sojae Reveal an Evolutionary Relationship between Oomycetes and Marine Vibrio Bacteria.
J.Biol.Chem., 286, 2011
|
|
4PCH
 
 | |
3O24
 
 | | Crystal structure of the brevianamide F prenyltransferase FtmPT1 from Aspergillus fumigatus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, CHLORIDE ION, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
3O2K
 
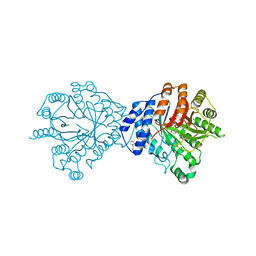 | | Crystal Structure of Brevianamide F Prenyltransferase Complexed with Brevianamide F and Dimethylallyl S-thiolodiphosphate | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
3LAT
 
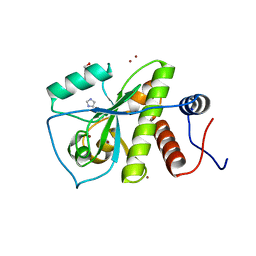 | |
3O8E
 
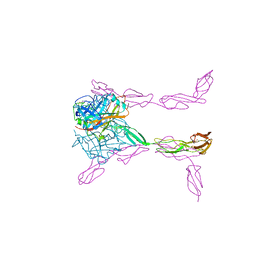 | | Structure of extracelllar portion of CD46 in complex with Adenovirus type 11 knob | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DITHIANE DIOL, Fiber 36.1 kDa protein, ... | | Authors: | Persson, B.D, Schmitz, N.B, Casasnovas, J.M, Stehle, T. | | Deposit date: | 2010-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens.
Plos Pathog., 6, 2010
|
|
4Y7T
 
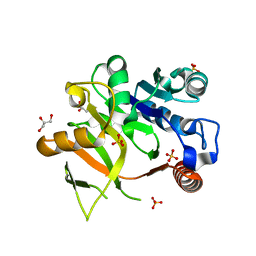 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7U
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7V
 
 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
3N0I
 
 | | Crystal Structure of Ad37 fiber knob in complex with GD1a oligosaccharide | | Descriptor: | Fiber, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ZINC ION | | Authors: | Nilsson, E.C, Storm, R.J, Bauer, J, Johansson, S.M.C, Lookene, A, Angstroem, J, Hedenstroem, M, Fraengsmyr, L, Rinaldi, S, Willison, H, Domelloef, F.P, Stehle, T, Arnberg, N. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis.
NAT.MED. (N.Y.), 17, 2011
|
|
3MK4
 
 | | X-Ray structure of human PEX3 in complex with a PEX19 derived peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Schmidt, F, Treiber, N, Dodt, G, Stehle, T. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19
J.Biol.Chem., 285, 2010
|
|
3PH5
 
 | |
3PH6
 
 | |
7A9C
 
 | |
7AJP
 
 | | Crystal Structure of Human Adenovirus 56 Fiber Knob | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Strebl, M, Mindler, K, Stehle, T. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.382018 Å) | | Cite: | Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7B27
 
 | |
6ZRZ
 
 | | Crystal structure of 5-dimethylallyl tryptophan synthase from Streptomyces coelicolor in complex with DMASPP and Trp | | Descriptor: | DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, TRYPTOPHAN | | Authors: | Ostertag, E, Broger, K, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
6ZRX
 
 | | Crystal structure of 6-dimethylallyltryptophan synthase from Micromonospora olivasterospora in complex with DMASPP and Trp | | Descriptor: | DI(HYDROXYETHYL)ETHER, DMATS type aromatic prenyltransferase, S-(3-methylbut-2-en-1-yl) trihydrogen thiodiphosphate, ... | | Authors: | Ostertag, E, Stehle, T, Zocher, G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Substrate and Catalytic Promiscuity of Tryptophan Prenyltransferases.
J.Mol.Biol., 433, 2020
|
|
