8R9T
 
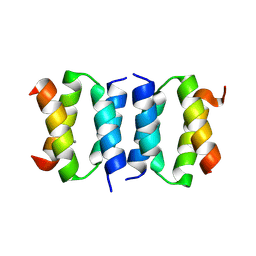 | |
8AXJ
 
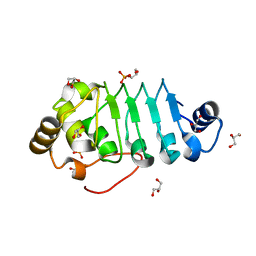 | |
8AXR
 
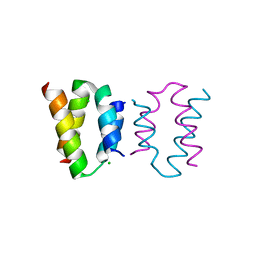 | | Crystal structure of the C-terminal domain of human CFAP410 | | Descriptor: | CHLORIDE ION, Cilia- and flagella-associated protein 410 | | Authors: | Dong, G, Stadler, A. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminus of CFAP410 forms a tetrameric helical bundle that is essential for its localization to the basal body.
Open Biology, 14, 2024
|
|
8AXO
 
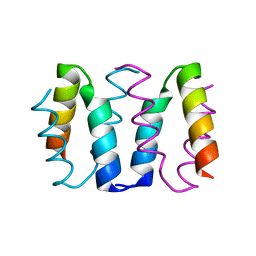 | |
4ZRG
 
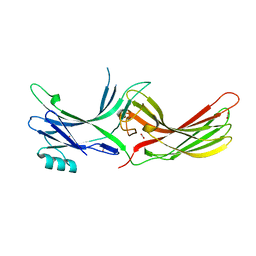 | | Visual arrestin mutant - R175E | | Descriptor: | CARBON DIOXIDE, S-arrestin | | Authors: | Granzin, J, Stadler, A, Cousin, A, Schlesinger, R, Batra-Safferling, R. | | Deposit date: | 2015-05-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the role of polar core residue Arg175 in arrestin activation.
Sci Rep, 5, 2015
|
|
4KUO
 
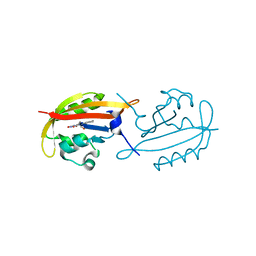 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4KUK
 
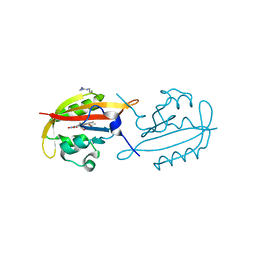 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
