4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-29 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5004 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
3P2A
 
 | | Crystal Structure of Thioredoxin 2 from Yersinia pestis | | 分子名称: | FORMIC ACID, Putative thioredoxin-like protein, ZINC ION | | 著者 | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-10-01 | | 公開日 | 2010-10-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.195 Å) | | 主引用文献 | Crystal Structure of Thioredoxin 2 from Yersinia pestis
TO BE PUBLISHED
|
|
3OT5
 
 | | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes | | 分子名称: | DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 2-epimerase | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-10 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 2.2 Angstrom Resolution Crystal Structure of putative UDP-N-acetylglucosamine 2-epimerase from Listeria monocytogenes.
TO BE PUBLISHED
|
|
3OZH
 
 | | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis | | 分子名称: | beta-lactamase/D-alanine carboxypeptidase | | 著者 | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-24 | | 公開日 | 2010-10-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.907 Å) | | 主引用文献 | Crystal Structure of Beta-Lactamase/D-alanine Carboxypeptidase from Yersinia pestis
To be Published
|
|
3G0M
 
 | | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2 | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cysteine desulfuration protein sufE, ... | | 著者 | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-01-28 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of cysteine desulfuration protein SufE from Salmonella typhimurium LT2
To be Published
|
|
4MZ1
 
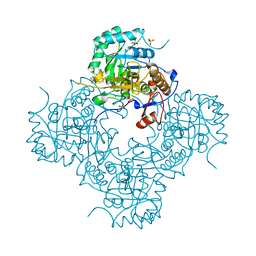 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | 分子名称: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-28 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3991 Å) | | 主引用文献 | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | 著者 | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-09-08 | | 公開日 | 2014-09-17 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8515 Å) | | 主引用文献 | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
4QVR
 
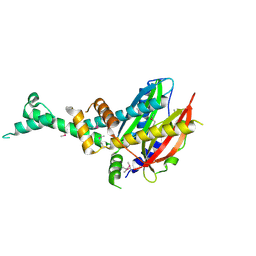 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | 分子名称: | Uncharacterized hypothetical protein FTT_1539c | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-07-15 | | 公開日 | 2014-07-30 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
3DR6
 
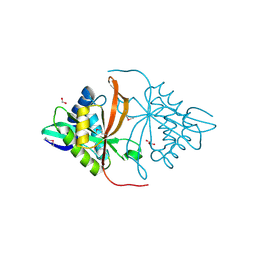 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | 著者 | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-07-10 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
4RS2
 
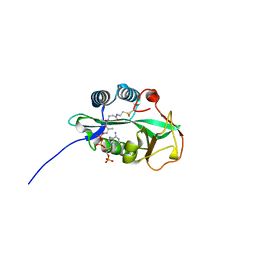 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | 分子名称: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | 著者 | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-11-06 | | 公開日 | 2014-11-19 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
4RN7
 
 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.717 Å) | | 主引用文献 | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
3OTR
 
 | | 2.75 Angstrom Crystal Structure of Enolase 1 from Toxoplasma gondii | | 分子名称: | CHLORIDE ION, Enolase, SULFATE ION | | 著者 | Minasov, G, Ruan, J, Shuvalova, L, Halavaty, A, Ngo, H, Tomavo, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-13 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The structure of bradyzoite-specific enolase from Toxoplasma gondii reveals insights into its dual cytoplasmic and nuclear functions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3OUU
 
 | | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni | | 分子名称: | Biotin carboxylase, CACODYLATE ION, CALCIUM ION, ... | | 著者 | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-15 | | 公開日 | 2010-10-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
4RO3
 
 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | 分子名称: | Hypothetical Protein, SULFATE ION | | 著者 | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-27 | | 公開日 | 2014-12-03 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
3EC6
 
 | | Crystal structure of the General Stress Protein 26 from Bacillus anthracis str. Sterne | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, General stress protein 26, SULFATE ION | | 著者 | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-08-29 | | 公開日 | 2008-09-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the general stress protein 26 from Bacillus anthracis str. Sterne
To be Published
|
|
4PUP
 
 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Burkholderia cenocepacia J2315 | | 分子名称: | Uncharacterized protein | | 著者 | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Minasov, G, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-03-13 | | 公開日 | 2014-04-16 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Burkholderia cenocepacia J2315
To be Published
|
|
4PZL
 
 | | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | Adenylate kinase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-03-31 | | 公開日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4PZ0
 
 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | 分子名称: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-03-28 | | 公開日 | 2014-04-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
3FWW
 
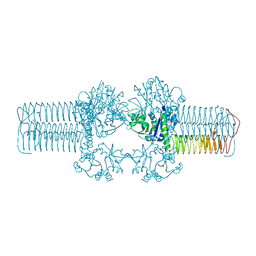 | | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92 | | 分子名称: | Bifunctional protein glmU | | 著者 | Zhang, R, Gu, M, Stam, J, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-01-19 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase from Yersinia pestis CO92
To be Published
|
|
3V4E
 
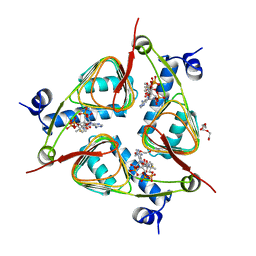 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | 分子名称: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | 著者 | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-12-14 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
4Q7G
 
 | | 1.7 Angstrom Crystal Structure of leukotoxin LukD from Staphylococcus aureus. | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leucotoxin LukDv | | 著者 | Minasov, G, Nocadello, S, Shuvalova, L, Shatsman, S, Kwon, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-04-24 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1FER
 
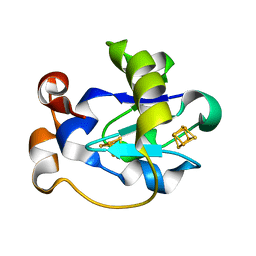 | | STRUCTURE AT PH 6.5 OF FERREDOXIN I FROM AZOTOBACTER VINELANDII AT 2.3 ANGSTROMS RESOLUTION | | 分子名称: | FE3-S4 CLUSTER, FERREDOXIN I, IRON/SULFUR CLUSTER | | 著者 | Merritt, E.A, Stout, G.H, Turley, S, Sieker, L.C, Jensen, L.H, Orme-Johnson, W.H. | | 登録日 | 1992-09-02 | | 公開日 | 1993-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure at pH 6.5 of ferredoxin I from Azotobacter vinelandii at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
3EFV
 
 | | Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative succinate-semialdehyde dehydrogenase | | 著者 | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-09-10 | | 公開日 | 2008-11-04 | | 最終更新日 | 2013-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | 分子名称: | Putative succinate-semialdehyde dehydrogenase | | 著者 | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-07 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3O2R
 
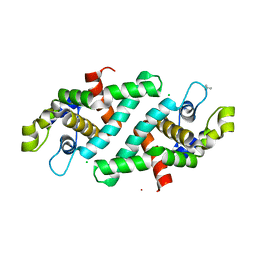 | | Structural flexibility in region involved in dimer formation of nuclease domain of Ribonuclase III (rnc) from Campylobacter jejuni | | 分子名称: | CHLORIDE ION, Ribonuclease III | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-07-22 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Structural Flexibility in Region Involved in Dimer Formation of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni.
TO BE PUBLISHED
|
|
