1LAJ
 
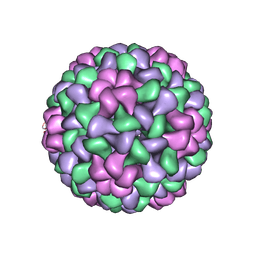 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | 分子名称: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | 登録日 | 2002-03-28 | | 公開日 | 2002-11-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
1O9V
 
 | | F17-aG lectin domain from Escherichia coli in complex with a selenium carbohydrate derivative | | 分子名称: | F17-AG LECTIN DOMAIN, methyl 2-acetamido-2-deoxy-1-seleno-beta-D-glucopyranoside | | 著者 | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | 登録日 | 2002-12-20 | | 公開日 | 2003-05-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1NUN
 
 | | Crystal Structure Analysis of the FGF10-FGFR2b Complex | | 分子名称: | Fibroblast growth factor-10, POLYETHYLENE GLYCOL (N=34), SULFATE ION, ... | | 著者 | Yeh, B.K, Igarashi, M, Eliseenkova, A.V, Plotnikov, A.N, Sher, I, Ron, D, Aaronson, S.A, Mohammadi, M. | | 登録日 | 2003-01-31 | | 公開日 | 2003-02-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis by which alternative splicing confers
specificity in fibroblast growth factor receptors.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2INY
 
 | |
1GQ1
 
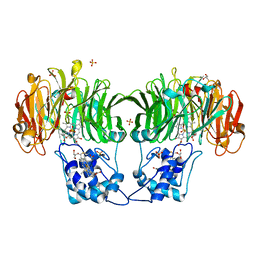 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | 分子名称: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | 著者 | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | 登録日 | 2001-11-19 | | 公開日 | 2002-11-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
5T6J
 
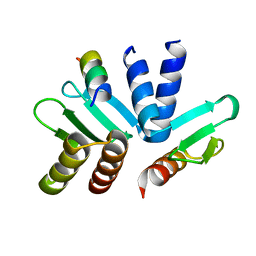 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | Kinetochore protein SPC24, Kinetochore protein SPC25, Kinetochore-associated protein DSN1 | | 著者 | Valverde, R, Jenni, S, Dimitrova, Y, Khin, Y, Harrison, S.C. | | 登録日 | 2016-09-01 | | 公開日 | 2016-11-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | 分子名称: | F17-AG LECTIN DOMAIN | | 著者 | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | 登録日 | 2002-12-23 | | 公開日 | 2003-05-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
5T51
 
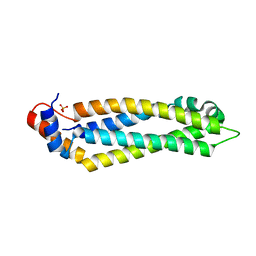 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | KLLA0E05809p, KLLA0F02343p, SULFATE ION | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2007 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
5T59
 
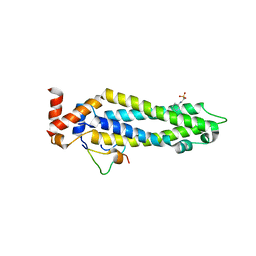 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, KLLA0B13629p, KLLA0E05809p, ... | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
1O8T
 
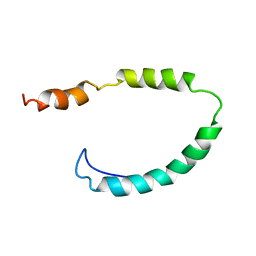 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | 分子名称: | APOLIPOPROTEIN C-II | | 著者 | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | 登録日 | 2002-11-29 | | 公開日 | 2003-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
5TD8
 
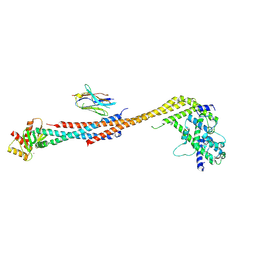 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | 分子名称: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | 著者 | Valverde, R, Ingram, J, Harrison, S.C. | | 登録日 | 2016-09-17 | | 公開日 | 2016-11-16 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (7.531 Å) | | 主引用文献 | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
1I85
 
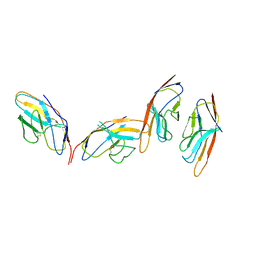 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | 分子名称: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | 著者 | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | 登録日 | 2001-03-12 | | 公開日 | 2001-04-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
1F3M
 
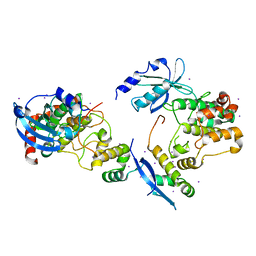 | | CRYSTAL STRUCTURE OF HUMAN SERINE/THREONINE KINASE PAK1 | | 分子名称: | IODIDE ION, SERINE/THREONINE-PROTEIN KINASE PAK-ALPHA | | 著者 | Lei, M, Lu, W, Meng, W, Parrini, M.-C, Eck, M.J, Mayer, B.J, Harrison, S.C. | | 登録日 | 2000-06-05 | | 公開日 | 2000-06-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch.
Cell(Cambridge,Mass.), 102, 2000
|
|
5T58
 
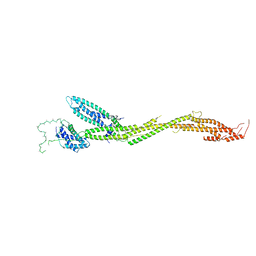 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | 分子名称: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | 著者 | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2131 Å) | | 主引用文献 | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
1PZU
 
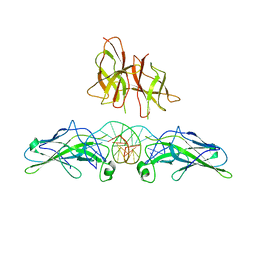 | | An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site | | 分子名称: | 5'-D(*AP*AP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3', 5'-D(*TP*TP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | 著者 | Jin, L, Sliz, P, Chen, L, Macian, F, Rao, A, Hogan, P.G, Harrison, S.C. | | 登録日 | 2003-07-14 | | 公開日 | 2003-09-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | An asymmetric NFAT1 dimer on a pseudo-palindromic KB-like DNA site
Nat.Struct.Biol., 10, 2003
|
|
1IC8
 
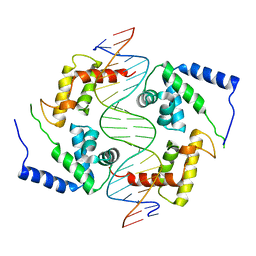 | | HEPATOCYTE NUCLEAR FACTOR 1A BOUND TO DNA : MODY3 GENE PRODUCT | | 分子名称: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*G)-3', HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | 著者 | Chi, Y.-I, Frantz, J.D, Oh, B.-C, Hansen, L, Dhe-Paganon, S, Shoelson, S.E. | | 登録日 | 2001-03-30 | | 公開日 | 2002-11-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Diabetes mutations delineate an
atypical POU domains in HNF1-Alpha
Mol.Cell, 10, 2002
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | 分子名称: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | 著者 | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | 登録日 | 2000-07-13 | | 公開日 | 2000-08-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
1BQI
 
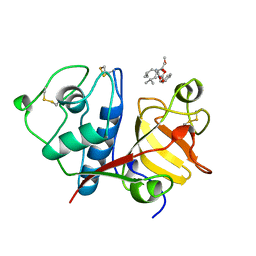 | | USE OF PAPAIN AS A MODEL FOR THE STRUCTURE-BASED DESIGN OF CATHEPSIN K INHIBITORS. CRYSTAL STRUCTURES OF TWO PAPAIN INHIBITOR COMPLEXES DEMONSTRATE BINDING TO S'-SUBSITES. | | 分子名称: | CARBOBENZYLOXY-(L)-LEUCINYL-(L)LEUCINYL METHOXYMETHYLKETONE, PAPAIN | | 著者 | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | 登録日 | 1998-08-16 | | 公開日 | 1999-08-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Use of papain as a model for the structure-based design of cathepsin K inhibitors: crystal structures of two papain-inhibitor complexes demonstrate binding to S'-subsites.
J.Med.Chem., 41, 1998
|
|
2KCX
 
 | |
1IK9
 
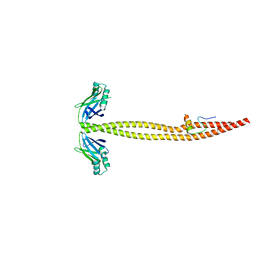 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | 分子名称: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | 著者 | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | 登録日 | 2001-05-03 | | 公開日 | 2001-11-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
5UK2
 
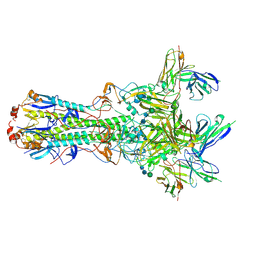 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UJZ
 
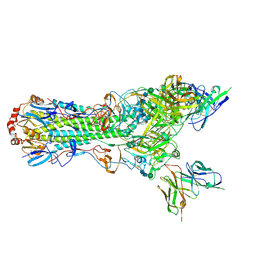 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
1IFR
 
 | |
5UG0
 
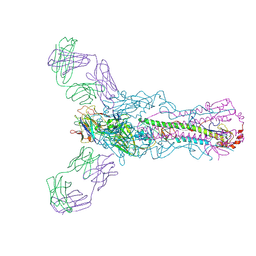 | | Human antibody H2897 in complex with influenza hemagglutinin H1 Solomon Islands/03/2006 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2897 heavy chain, ... | | 著者 | Raymond, D.D, Caradonna, T, Schmidt, A.G, Harrison, S.C. | | 登録日 | 2017-01-06 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UK0
 
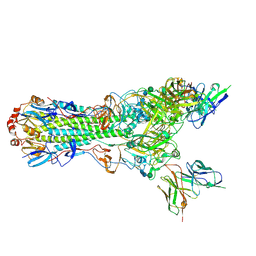 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
