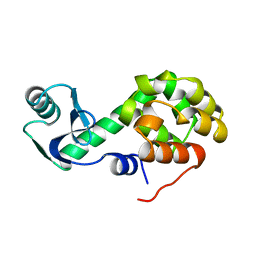
1L21
 
 | |
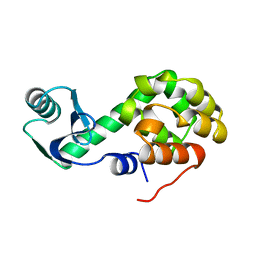
1L33
 
 | |
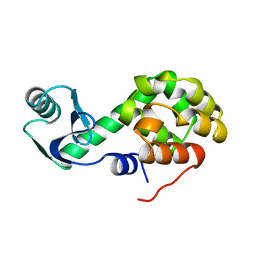
1L24
 
 | |
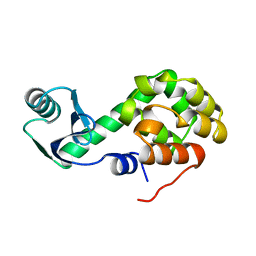
1L19
 
 | |
1L22
 
 | |
1L23
 
 | |
1L20
 
 | |
4EML
 
 | |
1AZP
 
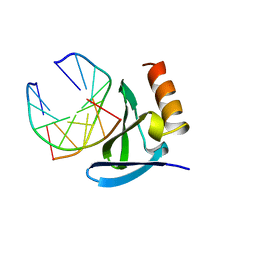 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
1AZQ
 
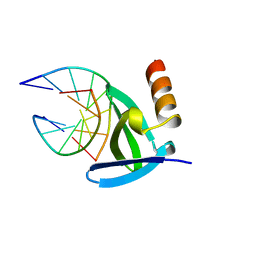 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
2XMP
 
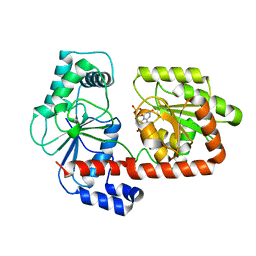 | | Crystal structure of trehalose synthase TreT mutant E326A from P. horishiki in complex with UDP | | Descriptor: | TREHALOSE-SYNTHASE TRET, URIDINE-5'-DIPHOSPHATE | | Authors: | Song, H.-N, Jung, T.-Y, Yoon, S.-M, Lee, S.-B, Lim, M.-Y, Woo, E.-J. | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights on the New Mechanism of Trehalose Synthesis by Trehalose Synthase Tret from Pyrococcus Horikoshii.
J.Mol.Biol., 404, 2010
|
|
5OUF
 
 | |
4I52
 
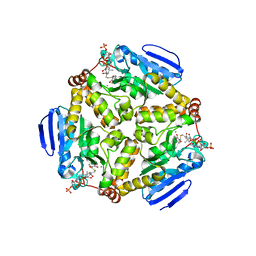 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I4Z
 
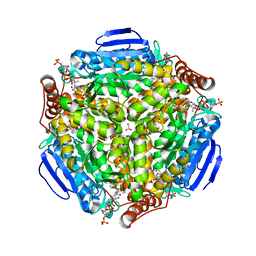 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4QII
 
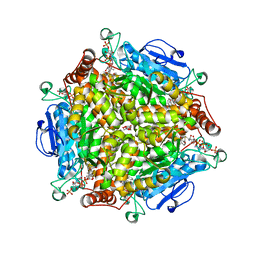 | | Crystal Structure of type II MenB from Mycobacteria tuberculosis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, Salicylyl CoA, TRIETHYLENE GLYCOL | | Authors: | Song, H.G, Tse, Y.S, Sung, H.P, Guo, Z.H. | | Deposit date: | 2014-05-31 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-dependent active-site closure revealed in the crystal structure of Mycobacterium tuberculosis MenB complexed with product analogues
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1XD2
 
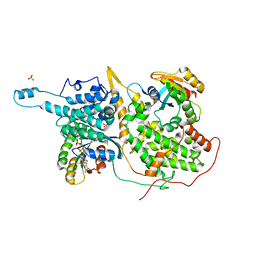 | | Crystal Structure of a ternary Ras:SOS:Ras*GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Sondermann, H, Soisson, S.M, Boykevisch, S, Yang, S.S, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2004-09-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of autoinhibition in the ras activator son of sevenless.
Cell(Cambridge,Mass.), 119, 2004
|
|
1OOW
 
 | | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f | | Descriptor: | COPPER (II) ION, Plastocyanin, chloroplast | | Authors: | Jansson, H, Okvist, M, Jacobson, F, Ejdeback, M, Hansson, O, Sjolin, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f.
Biochim.Biophys.Acta, 1607, 2003
|
|
1AL9
 
 | |
1AMD
 
 | |
1AWX
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
1B5D
 
 | | DCMP Hydroxymethylase from T4 (Intact) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B49
 
 | | DCMP HYDROXYMETHYLASE FROM T4 (PHOSPHATE-BOUND) | | Descriptor: | PHOSPHATE ION, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1BHR
 
 | | 2'-DEOXY-ISOGUANOSINE BASE PAIRED TO THYMIDINE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*IGUP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
1AWW
 
 | | SH3 DOMAIN FROM BRUTON'S TYROSINE KINASE, NMR, 42 STRUCTURES | | Descriptor: | BRUTON'S TYROSINE KINASE | | Authors: | Hansson, H, Mattsson, P.T, Allard, P, Haapaniemi, P, Vihinen, M, Smith, C.I.E, Hard, T. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from Bruton's tyrosine kinase.
Biochemistry, 37, 1998
|
|
4KKQ
 
 | |
