6IUN
 
 | |
6IUM
 
 | |
8JB1
 
 | |
8HRP
 
 | |
8HRR
 
 | |
8HRT
 
 | |
8HRQ
 
 | |
8HRS
 
 | |
5X5U
 
 | |
8HRO
 
 | |
5X5T
 
 | |
3ZHK
 
 | |
3ZHD
 
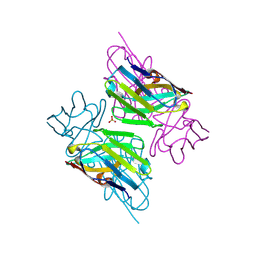 | |
3ZHL
 
 | |
5X7N
 
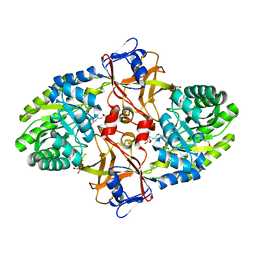 | |
5VAG
 
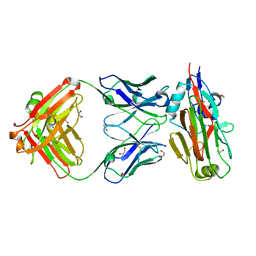 | | Crystal structure of H7-specific antibody m826 in complex with the HA1 domain of hemagglutinin from H7N9 influenza virus | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody m826, Hemagglutinin, ... | | Authors: | Song, H, Ying, T, Ji, X. | | Deposit date: | 2017-03-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.
Cell Host Microbe, 22, 2017
|
|
2VR5
 
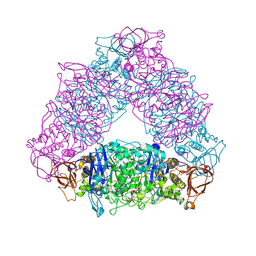 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
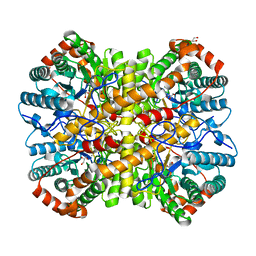
5YN3
 
 | |
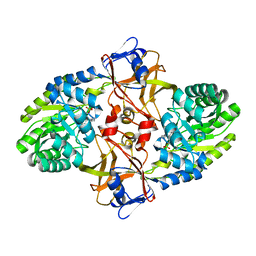
5X7M
 
 | |
4URP
 
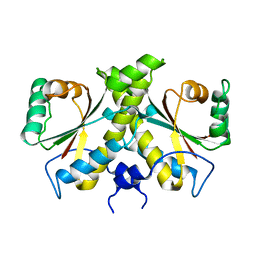 | | The Crystal structure of Nitroreductase from Saccharomyces cerevisiae | | Descriptor: | FATTY ACID REPRESSION MUTANT PROTEIN 2 | | Authors: | Song, H.-N, Woo, E.-J, Bang, S.-Y, Jung, D.-G, Park, S.-G. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Crystal Structure of the Fungal Nitroreductase Frm2 from Saccharomyces Cerevisiae.
Protein Sci., 24, 2015
|
|
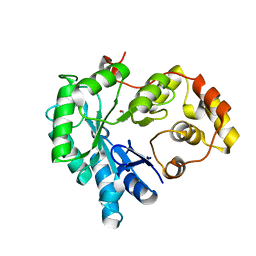
5Z6U
 
 | |
5Z6T
 
 | |
5IJZ
 
 | |
5HXX
 
 | | Crystal structure of AspAT from Corynebacterium glutamicum | | Descriptor: | 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, ... | | Authors: | Son, H.-F, Kim, K.-J. | | Deposit date: | 2016-01-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into a novel class of aspartate aminotransferase from Corynebacterium glutamicum
To be published
|
|
5IWQ
 
 | |
