7QHV
 
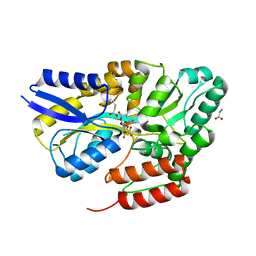 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with sulfoquinovosyl diacylglycerol | | Descriptor: | GLYCINE, Sulfoquinovosyl binding protein, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-[(2~{S})-3-butanoyloxy-2-heptanoyloxy-propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A, Davies, G.J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7NBZ
 
 | |
7YZS
 
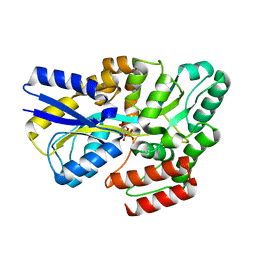 | |
7YZU
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with SQMe | | Descriptor: | Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-methoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Davies, G.J. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7BBZ
 
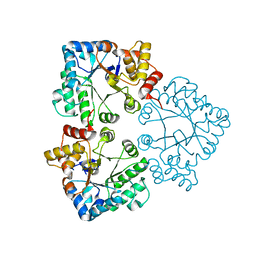 | |
7BC0
 
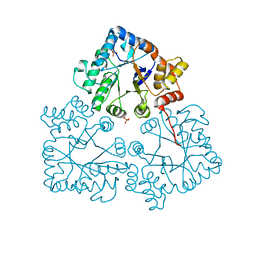 | |
7BBY
 
 | |
7BC1
 
 | |
8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8C4I
 
 | |
8S5B
 
 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
1I1R
 
 | | CRYSTAL STRUCTURE OF A CYTOKINE/RECEPTOR COMPLEX | | Descriptor: | INTERLEUKIN-6 RECEPTOR BETA CHAIN, VIRAL IL-6 | | Authors: | Chow, D, He, X, Snow, A.L, Rose-John, S, Garcia, K.C. | | Deposit date: | 2001-02-02 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an extracellular gp130 cytokine receptor signaling complex.
Science, 291, 2001
|
|
1PXO
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2-Amino-4-methyl-thiazol-5-yl)-pyrimidin-2-yl]-(3-nitro-phenyl)-amine | | Descriptor: | Cell division protein kinase 2, [4-(2-AMINO-4-METHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-(3-NITRO-PHENYL)-AMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXM
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 3-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 3-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXN
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-[4-(4-Methyl-2-methylamino-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 4-[4-(4-METHYL-2-METHYLAMINO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PXP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-N',N'-dimethyl-benzene-1,4-diamine | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-THIAZOL-5-YL)-PYRIMIDIN-2-YL]-N',N'-DIMETHYL-BENZENE-1,4-DIAMINE | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
