4NCC
 
 | | Neutralizing antibody to murine norovirus | | Descriptor: | Fab fragment heavy, Fab fragment light | | Authors: | Smith, T, Li, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization.
J.Virol., 88, 2014
|
|
4GVB
 
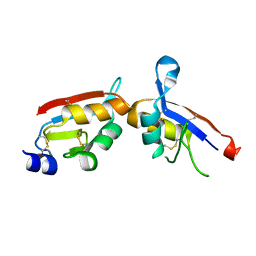 | |
4LLF
 
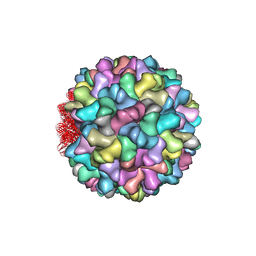 | | Crystal structure of Cucumber Necrosis Virus | | Descriptor: | CALCIUM ION, Capsid protein, ZINC ION | | Authors: | Smith, T. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8891 Å) | | Cite: | Atomic structure of cucumber necrosis virus and the role of the capsid in vector transmission.
J.Virol., 87, 2013
|
|
6EEM
 
 | | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | | Descriptor: | N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-tyrosine, SULFATE ION, TYROSINE, ... | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61000657 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEQ
 
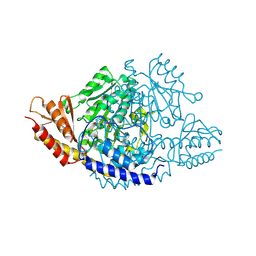 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | Descriptor: | 4-hydroxyphenylacetaldehyde synthase | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.600086 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEI
 
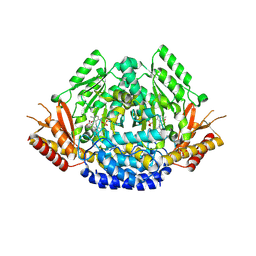 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | Descriptor: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-14 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (1.99001348 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEW
 
 | | Crystal structure of Catharanthus roseus tryptophan decarboxylase in complex with L-tryptophan | | Descriptor: | Aromatic-L-amino-acid decarboxylase, CALCIUM ION, TRYPTOPHAN | | Authors: | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | Deposit date: | 2018-08-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.05002069 Å) | | Cite: | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4EO7
 
 | | Crystal structure of the TIR domain of human myeloid differentiation primary response protein 88. | | Descriptor: | MAGNESIUM ION, Myeloid differentiation primary response protein MyD88 | | Authors: | Snyder, G.A, Cirl, C, Jiang, J.S, Chen, P, Smith, T, Xiao, T.S. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7T8J
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a GSGG linker | | Descriptor: | CHLORIDE ION, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Soleimani, S, Pedroza Romo, M.J, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
1KPT
 
 | |
7U4W
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at traditional protein concentration | | Descriptor: | Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Pedroza Romo, M.J, Smith, T, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
8FZ4
 
 | | The von Willebrand factor A domain of Anthrax toxin receptor 2 | | Descriptor: | Anthrax toxin receptor 2, CHLORIDE ION, MAGNESIUM ION | | Authors: | Pedroza-Romo, M.J, Soleimani, S, Lewis, A, Smith, T, Brown, S, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7U4Z
 
 | |
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
