6DNL
 
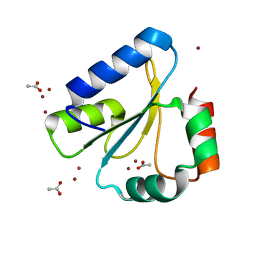 | | Crystal Structure of Neisseria meningitidis DsbD c-terminal domain in the reduced form | | Descriptor: | ACETATE ION, Thiol:disulfide interchange protein DsbD, ZINC ION | | Authors: | Smith, R.P, Heras, B, Paxman, J.J. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the disulfide reductase mechanism of DsbD, an essential enzyme for neisserial pathogens.
J. Biol. Chem., 293, 2018
|
|
1SRB
 
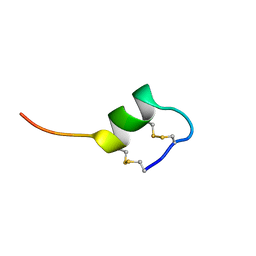 | | CONFORMATIONAL STUDIES ON SRTB, A NON-SELECTIVE ENDOTHELIN RECEPTOR AGONIST, AND ON IRL 1620, AN ETB RECEPTOR SPECIFIC AGONIST | | Descriptor: | Sarafotoxins precursor | | Authors: | Smith, R, Atkins, A.R. | | Deposit date: | 1994-09-06 | | Release date: | 1994-11-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | 1H NMR studies of sarafotoxin SRTb, a nonselective endothelin receptor agonist, and IRL 1620, an ETB receptor-specific agonist.
Biochemistry, 34, 1995
|
|
6DNV
 
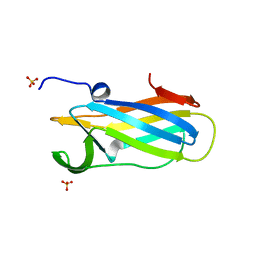 | |
1TES
 
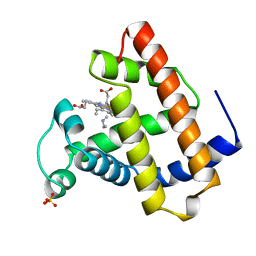 | | OXYGEN BINDING MUSCLE PROTEIN | | Descriptor: | METHYLETHYLAMINE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Phillips Jr, G.N, Olson, J.S. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of NO-induced oxidation of myoglobin and hemoglobin.
Biochemistry, 35, 1996
|
|
101M
 
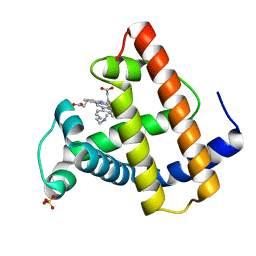 | | SPERM WHALE MYOGLOBIN F46V N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-13 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
102M
 
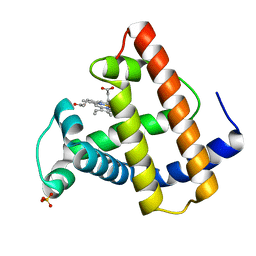 | | SPERM WHALE MYOGLOBIN H64A AQUOMET AT PH 9.0 | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-15 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
103M
 
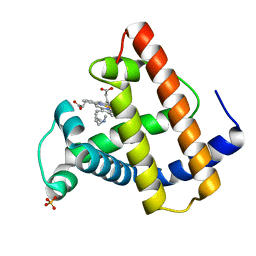 | | SPERM WHALE MYOGLOBIN H64A N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
105M
 
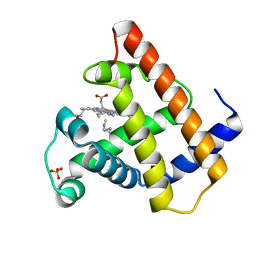 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
106M
 
 | | SPERM WHALE MYOGLOBIN V68F ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-21 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
104M
 
 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
107M
 
 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
108M
 
 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-23 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
109M
 
 | | SPERM WHALE MYOGLOBIN D122N ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
110M
 
 | | SPERM WHALE MYOGLOBIN D122N METHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | METHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
111M
 
 | | SPERM WHALE MYOGLOBIN D122N N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-24 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
112M
 
 | | SPERM WHALE MYOGLOBIN D122N N-PROPYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-PROPYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-24 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
1MIH
 
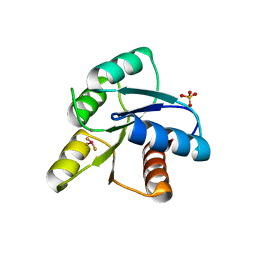 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
6DNU
 
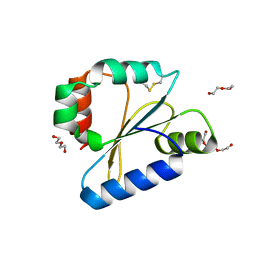 | |
7YJL
 
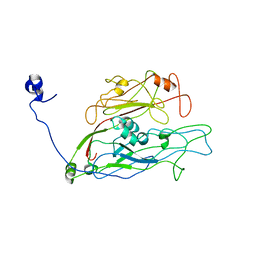 | |
4PED
 
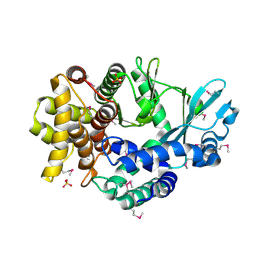 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
1PLP
 
 | | SOLUTION STRUCTURE OF THE CYTOPLASMIC DOMAIN OF PHOSPHOLAMBAN | | Descriptor: | PHOSPHOLAMBAN | | Authors: | Mortishire-Smith, R.J, Pitzenberger, S.M, Burke, C.J, Middaugh, C.R, Garsky, V.M, Johnson, R.G. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytoplasmic domain of phopholamban: phosphorylation leads to a local perturbation in secondary structure.
Biochemistry, 34, 1995
|
|
1POU
 
 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
1OJY
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK2
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK3
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
