4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | Descriptor: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | Authors: | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
1KXZ
 
 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3I
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3C
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP WITH SHORT CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of Mt0146/Cbit Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
4MDE
 
 | | Structure of bacterial polynucleotide kinase product complex bound to GDP and DNA | | Descriptor: | DNA (5'-D(P*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
4MDF
 
 | | Structure of bacterial polynucleotide kinase Michaelis complex bound to GTP and DNA | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*TP*GP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shuman, S, Das, U, Wang, L.K, Smith, P, Jacewicz, A. | | Deposit date: | 2013-08-22 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5'-OH oligonucleotide and a product complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition.
Nucleic Acids Res., 42, 2014
|
|
4IFP
 
 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
4IKM
 
 | | X-ray structure of CARD8 CARD domain | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Jin, T, Huang, M, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4606 Å) | | Cite: | The structure of the CARD8 caspase-recruitment domain suggests its association with the FIIND domain and procaspases through adjacent surfaces.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IRL
 
 | | X-ray structure of the CARD domain of zebrafish GBP-NLRP1 like protein | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jin, T, Huang, M, Smith, P, Xiao, T. | | Deposit date: | 2013-01-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of the caspase-recruitment domain from a zebrafish guanylate-binding protein.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JST
 
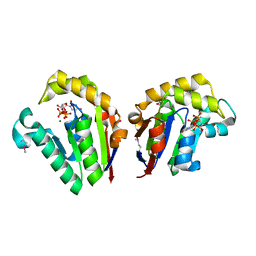 | | Structure of Clostridium thermocellum polynucleotide kinase bound to UTP | | Descriptor: | MAGNESIUM ION, Metallophosphoesterase, SODIUM ION, ... | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
4JT4
 
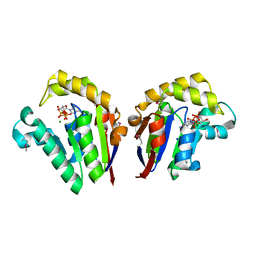 | | Structure of Clostridium thermocellum polynucleotide kinase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase, ... | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
4JT2
 
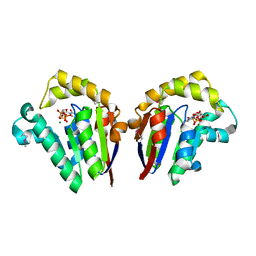 | | Structure of Clostridium thermocellum polynucleotide kinase bound to CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
1NKV
 
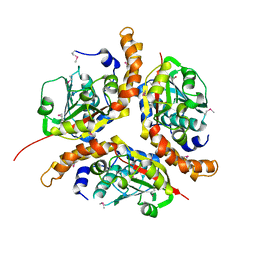 | | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13 | | Descriptor: | HYPOTHETICAL PROTEIN yjhP | | Authors: | Kuzin, A, Manor, P, Benach, J, Smith, P, Rost, B, Xiao, R, Montelione, G, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-03 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-RAY STRUCTURE OF YJHP FROM E.COLI NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER13
To be published
|
|
4J6O
 
 | | Crystal Structure of the Phosphatase Domain of C. thermocellum (Bacterial) PnkP | | Descriptor: | CITRIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wang, L, Smith, P, Shuman, S. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the 2',3' phosphatase component of the bacterial Pnkp-Hen1 RNA repair system.
Nucleic Acids Res., 41, 2013
|
|
4JSY
 
 | | Structure of Clostridium thermocellum polynucleotide kinase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Metallophosphoesterase | | Authors: | Das, U, Wang, L.K, Smith, P, Shuman, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkphen1 RNA repair system.
Biochemistry, 52, 2013
|
|
1N4A
 
 | | The Ligand Bound Structure of E.coli BtuF, the Periplasmic Binding Protein for Vitamin B12 | | Descriptor: | CYANOCOBALAMIN, Vitamin B12 transport protein btuF | | Authors: | Karpowich, N.K, Smith, P.C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-30 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding.
J.Biol.Chem., 278, 2003
|
|
1N4D
 
 | |
1MZH
 
 | | QR15, an Aldolase | | Descriptor: | Deoxyribose-phosphate aldolase, PHOSPHATE ION | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aquifex Aeolicus Aldolase,
Northeast Structural Genomics Consortium Target
QR15
To be Published
|
|
1M1S
 
 | | Structure of WR4, a C.elegans MSP family member | | Descriptor: | WR4 | | Authors: | Karpowich, N, Smith, P, Shen, J, Hunt, J, Montelione, G, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-20 | | Release date: | 2003-07-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a C.elegans MSP family member
To be Published
|
|
1F38
 
 | |
5FQS
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 3. | | Descriptor: | (E)-3-[4-(6-HYDROXY-2-ISOBUTYL-1-METHYL-3,4-DIHYDROISOQUINOLIN-1-YL)PHENYL]PROP-2-ENOIC ACID, ESTROGEN RECEPTOR | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
5FQP
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 1. | | Descriptor: | (E)-3-[4-[(1R,3R)-6-hydroxy-2-isobutyl-3-methyl-3,4-dihydro-1H-isoquinolin-1-yl]phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
5FQR
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 2. | | Descriptor: | (E)-3-[4-[(1R)-6-HYDROXY-2-ISOBUTYL-3,4-DIHYDRO-1H-ISOQUINOLIN-1-YL]PHENYL]PROP-2-ENOIC ACID, ESTROGEN RECEPTOR | | Authors: | Scott, J.S, Bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
5FQV
 
 | | Selective estrogen receptor downregulator antagonists: Tetrahydroisoquinoline phenols 5. | | Descriptor: | (E)-3-[4-(6-hydroxy-2-isobutyl-7-methyl-3,4-dihydro-1H-isoquinolin-1-yl)phenyl]prop-2-enoic acid, ESTROGEN RECEPTOR ALPHA | | Authors: | Scott, J.S, bailey, A, Davies, R.D.M, Degorce, S.L, MacFaul, P.A, Gingell, H, Moss, T, Norman, R.A, Pink, J.H, Rabow, A.A, Roberts, B, Smith, P.D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tetrahydroisoquinoline Phenols: Selective Estrogen Receptor Downregulator Antagonists with Oral Bioavailability in Rat.
Acs Med.Chem.Lett., 7, 2016
|
|
