7NII
 
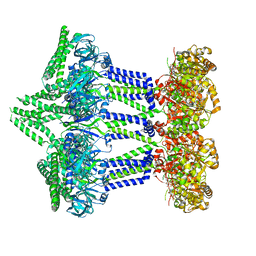 | | Wzc-K540M MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
1QD9
 
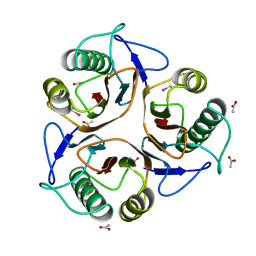 | | Bacillus subtilis YABJ | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, MERCURY (II) ION, ... | | Authors: | Smith, J.L, Sinha, S, Rappu, P, Lange, S.C, Mantsala, P, Zalkin, H. | | Deposit date: | 1999-07-09 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7NHS
 
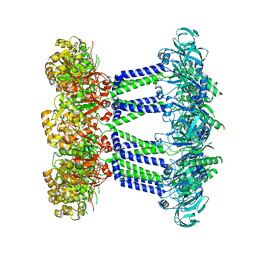 | | Wzc K540M C8 | | Descriptor: | Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NI2
 
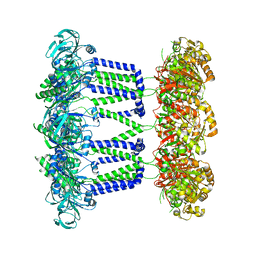 | | Wzc-K540M-4YE C8 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NIB
 
 | | Wzc-K540M-4YE C1 | | Descriptor: | Tyrosine-protein kinase | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
1DSM
 
 | | (-)-duocarmycin SA covalently linked to duplex DNA | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, 5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3' | | Authors: | Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structural basis for in situ activation of DNA alkylation by duocarmycin SA
J.Mol.Biol., 300, 2000
|
|
5FFM
 
 | | Yellow fever virus helicase | | Descriptor: | Serine protease NS3 | | Authors: | Smith, J.L. | | Deposit date: | 2015-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing.
J. Virol., 79, 2005
|
|
1GPH
 
 | |
7N2T
 
 | | O-acetylserine sulfhydrylase from Citrullus vulgaris in the internal aldimine state, with citrate bound | | Descriptor: | CITRIC ACID, Cysteine synthase, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, J.L, Buller, A.R, Bingman, C.A. | | Deposit date: | 2021-05-29 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of beta-Substitution Activity of O-Acetylserine Sulfhydrolase from Citrullus vulgaris.
Chembiochem, 23, 2022
|
|
1D0T
 
 | |
6YZ5
 
 | | H11-D4 complex with SARS-CoV-2 RBD | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, Le Bas, A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
6Z3C
 
 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z3B
 
 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6YQQ
 
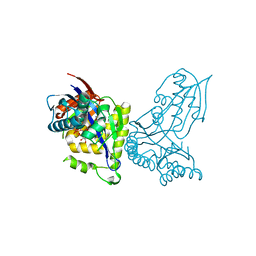 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
6Y47
 
 | |
1D0U
 
 | |
6YZ7
 
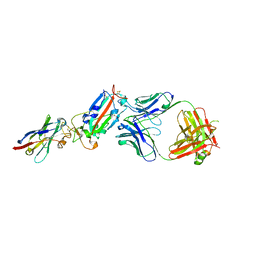 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
6Z2M
 
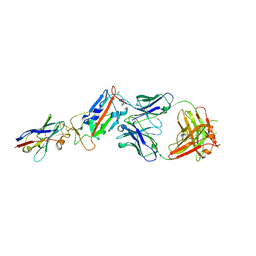 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
6Z33
 
 | |
6Z2N
 
 | |
6YY5
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with TCV_L5 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[2-[[(2~{S})-3-[[(2~{S})-3-[[1-[2-[2-[2-[4-[4-[5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazin-1-yl]-2-oxidanylidene-ethoxy]ethoxy]ethyl]-1,2,3-triazol-4-yl]methylamino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Naismith, J.H, Moynie, L.M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa.
Acs Infect Dis., 2022
|
|
7S3N
 
 | |
7S3M
 
 | |
5N0O
 
 | |
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
