3EBW
 
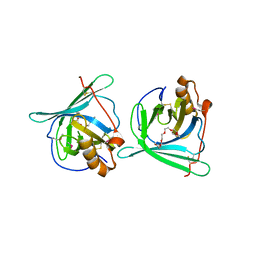 | | Crystal structure of major allergens, Per a 4 from cockroaches | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Per a 4 allergen | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
3E3C
 
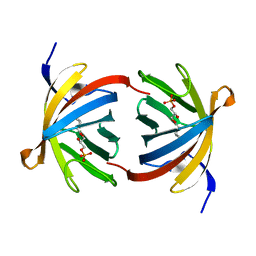 | | Structure of GrlR-lipid complex | | Descriptor: | (2R)-2-HYDROXY-3-(PHOSPHONOOXY)PROPYL HEXANOATE, L0044 | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2008-08-07 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of the lipid binding property of GrlR, a locus of enterocyte effacement regulator.
Biochem.J., 2009
|
|
3H8B
 
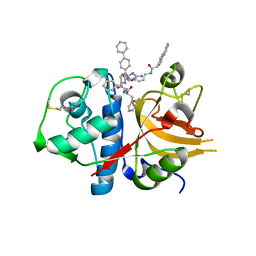 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3H89
 
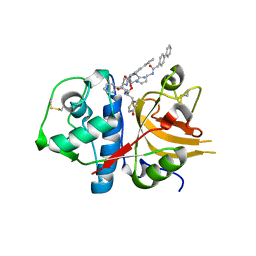 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 4) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3H8C
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors (compound 14) | | Descriptor: | Cathepsin L1, N-(biphenyl-4-ylacetyl)-S-methyl-L-cysteinyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Combined Crystallographic and Molecular Dynamics Study of Cathepsin L Retrobinding Inhibitors
J.Med.Chem., 2009
|
|
3HH7
 
 | | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra) | | Descriptor: | Muscarinic toxin-like protein 3 homolog | | Authors: | Roy, A, Zhou, X, Chong, M.Z, D'hoedt, D, Foo, C.S, Rajagopalan, N, Nirthanan, S, Bertrand, D, Sivaraman, J, Kini, R.M. | | Deposit date: | 2009-05-15 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Functional Characterization of a Novel Homodimeric Three-finger Neurotoxin from the Venom of Ophiophagus hannah (King Cobra)
J.Biol.Chem., 285, 2010
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3T7S
 
 | |
3SXJ
 
 | |
3T0I
 
 | |
2MQ1
 
 | | Phosphotyrosine binding domain | | Descriptor: | E3 ubiquitin-protein ligase Hakai, ZINC ION | | Authors: | Mukherjee, M, Jing-Song, F, Sivaraman, J. | | Deposit date: | 2014-06-11 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dimeric switch of Hakai-truncated monomers during substrate recognition: insights from solution studies and NMR structure.
J.Biol.Chem., 289, 2014
|
|
3T7T
 
 | |
3T7R
 
 | |
3SVZ
 
 | |
2Q1K
 
 | | Cyrstal Structure of AscE from Aeromonas hydrophilla | | Descriptor: | AscE | | Authors: | Tan, Y.W, Yu, H.B, Leung, K.Y, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2007-05-24 | | Release date: | 2008-06-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila.
Protein Sci., 17, 2008
|
|
2QHD
 
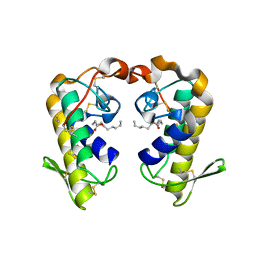 | | Crystal structure of ecarpholin S (ser49-PLA2) complexed with fatty acid | | Descriptor: | LAURIC ACID, Phospholipase A2 | | Authors: | Zhou, X, Tan, T.C, Valiyaveettil, S, Go, M.L, Kini, R.M, Sivaraman, J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
2QHE
 
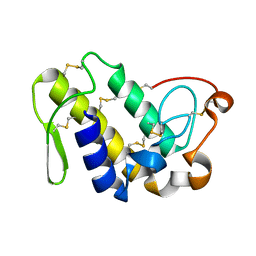 | | Crystal structure of Ser49-PLA2 (ecarpholin S) from Echis carinatus sochureki snake venom | | Descriptor: | Phospholipase A2 | | Authors: | Zhou, X, Valiyaveettil, S, Go, M.L, Kini, R.M, Sivaraman, J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
2OVS
 
 | |
2ZR5
 
 | |
2ZR4
 
 | |
2ZQT
 
 | |
2ZR6
 
 | |
2ZQS
 
 | |
3BJW
 
 | | Crystal Structure of ecarpholin S complexed with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Phospholipase A2 | | Authors: | Zhou, X, Sivaraman, J. | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Myotoxic Ecarpholin S from Echis carinatus Venom
Biophys.J., 95, 2008
|
|
2ZQU
 
 | |
