7PCK
 
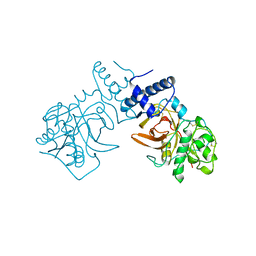 | |
1ZPS
 
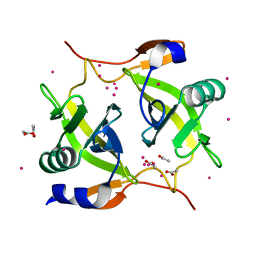 | | Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI | | Descriptor: | ACETIC ACID, CADMIUM ION, Phosphoribosyl-AMP cyclohydrolase | | Authors: | Sivaraman, J, Myers, R.S, Boju, L, Sulea, T, Cygler, M, Davisson, V.J, Schrag, J.D, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Methanobacterium thermoautotrophicum Phosphoribosyl-AMP Cyclohydrolase HisI.
Biochemistry, 44, 2005
|
|
5JYO
 
 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | Descriptor: | 2-(pyridin-2-yl)-N-(5-{4-[6-({[3-(trifluoromethoxy)phenyl]acetyl}amino)pyridazin-3-yl]butyl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Sivaraman, J, Jayaraman, S. | | Deposit date: | 2016-05-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
5DIN
 
 | |
3LCV
 
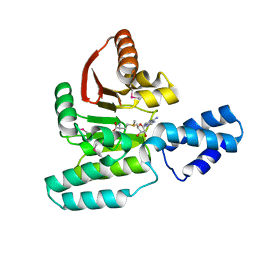 | | Crystal Structure of Antibiotic related Methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, Sisomicin-gentamicin resistance methylase Sgm | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-01-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases.
Nucleic Acids Res., 2010
|
|
3LCU
 
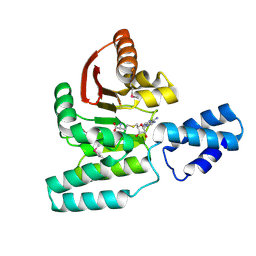 | | Crystal Structure of Antibiotic related Methyltransferase | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Sisomicin-gentamicin resistance methylase Sgm | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-01-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases.
Nucleic Acids Res., 2010
|
|
2R66
 
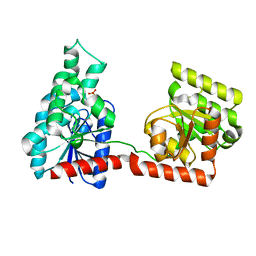 | |
2R68
 
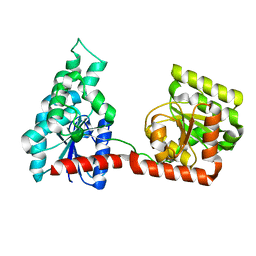 | |
2R60
 
 | |
5IAO
 
 | |
6K2C
 
 | | Extended Hect domain of UBE3C E3 Ligase | | Descriptor: | Ubiquitin-protein ligase E3C | | Authors: | Sivaraman, J, Singh, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of HECT domain of UBE3C E3 ligase and its ubiquitination activity.
Biochem.J., 477, 2020
|
|
6JX5
 
 | | Hect domain of AREL1 | | Descriptor: | Apoptosis-resistant E3 ubiquitin protein ligase 1 | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6JX6
 
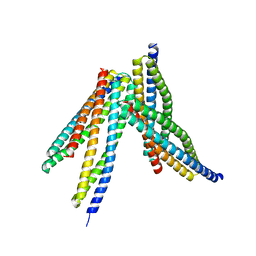 | | Tetrameric form of Smac | | Descriptor: | Diablo homolog, mitochondrial | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
1KSK
 
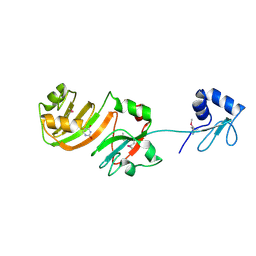 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1IJI
 
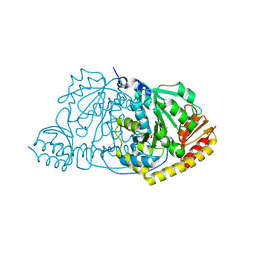 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1KSL
 
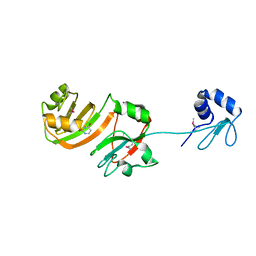 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1KSV
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URIDINE-5'-MONOPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-14 | | Release date: | 2002-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
1FG7
 
 | |
1FG3
 
 | | CRYSTAL STRUCTURE OF L-HISTIDINOL PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH L-HISTIDINOL | | Descriptor: | HISTIDINOL PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Cygler, M. | | Deposit date: | 2000-07-27 | | Release date: | 2001-08-22 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1PS7
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1DEU
 
 | | CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN X: A CYSTEINE PROTEASE WITH THE PROREGION COVALENTLY LINKED TO THE ACTIVE SITE CYSTEINE | | Descriptor: | PROCATHEPSIN X | | Authors: | Sivaraman, J, Nagler, D.K, Zhang, R, Menard, R, Cygler, M. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine.
J.Mol.Biol., 295, 2000
|
|
1PS6
 
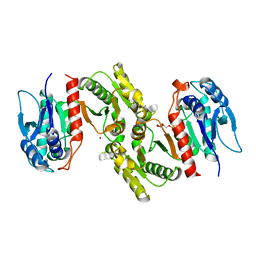 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1PRZ
 
 | | Crystal structure of pseudouridine synthase RluD catalytic module | | Descriptor: | Ribosomal large subunit pseudouridine synthase D | | Authors: | Sivaraman, J, Iannuzzi, P, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the RluD pseudouridine Synthase catalytic module, an
enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli
J.Mol.Biol., 335, 2003
|
|
1PTM
 
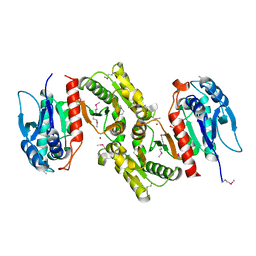 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1MC3
 
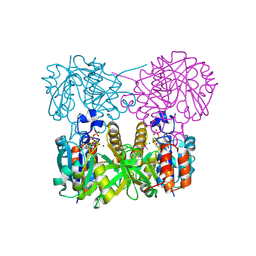 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
